The Learning Set was exported as an R workspace and transferred to cloud computing infrastructure for training. PMOD can be installed on the virtual machine provided by the cloud computing provider and training launched using the R workspace.
The parameters used were:
•AWS p2.8xlarge, 8 GPU K80, memory 64 GB
•382 samples (288 training, 72 validation, 22 testing)
•batch size 24
•Learning rate 0.005
•epochs 300
Training took 268 minutes at 0.07 seconds/sample. The loss values for training and validation were extracted from the Manifest and plotted:
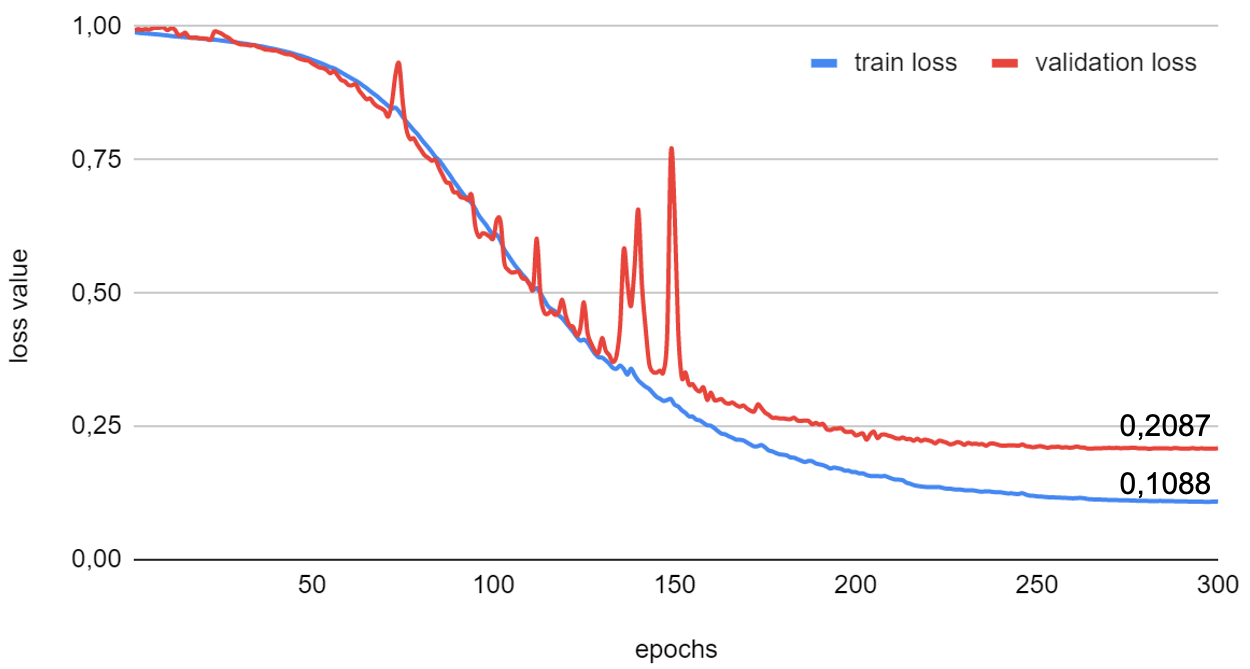
The plot illustrates how the loss value reduces with each epoch until a plateau is reached.
The Learning Set, Weights and Manifest were exported from the virtual machine and used to create a new model folder Rat Brain Dopaminergic PET in the weights subfolder of the Multichannel Segmentation folder in the PMOD installation /resources/pai folder. After this Deployment it was possible to test the model performance of comparable rat brain [11C]-methylphenidate data for which an anatomical MR image was available.
The resulting VOIs are shown below on the EARLY, LATE and anatomical reference MR images:
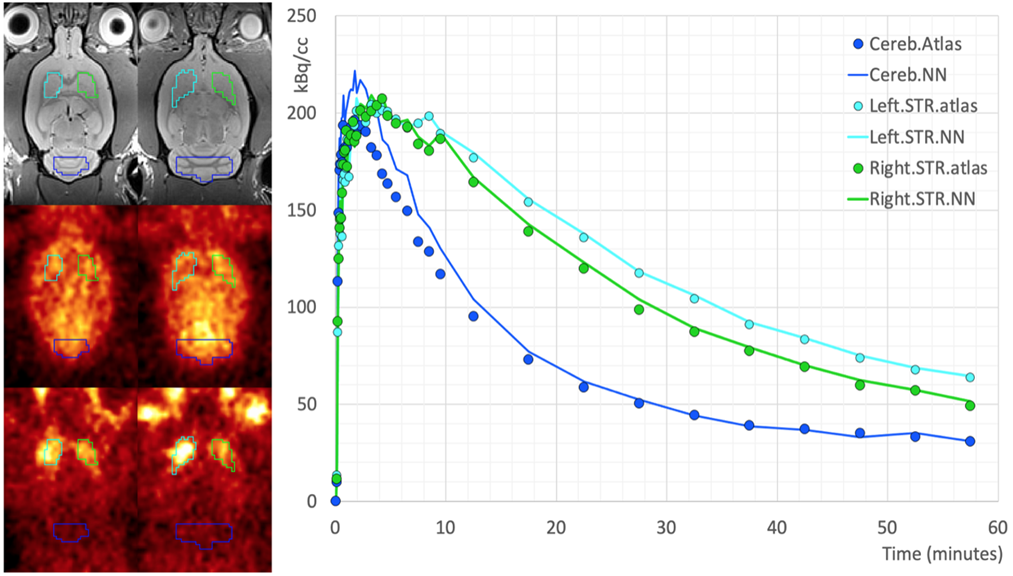
Time-activity curves were extracted from 8 dynamic PET series (4 RAC, 4 MP) using both sets of VOIs (AI-based, PNROD reference) and BPnd calculated using the SRTM model. The mean +/- SD BPnd using the AI-based VOIs was 1.29 +/- 0.67 and for PNROD VOIs 1.39 +/- 0.79.
Example data to test the Rat Brain Dopaminergic PET model is available in our Demo database (Subject PAI5). The data used for the entire case study was kindly provided by Prof. Kristina Herfert at the Werner Siemens Imaging Center at the University of Tuebingen in Germany.
To try the model for yourself we recommend use of the Segment tool (PSEG). Select the early average series as Input series. The late average series will be detected automatically according to the Association mechanism. The model selection must be Rat Brain Dopaminergic PET (Multichannel Segmentation). The model was trained with 3D data so no Split Slices/Frames is available or required. The resulting VOIs can be saved and used to extract TACs from the Inveon dynamic PET series (e.g. standalone analysis in View, or multimodal analysis combined with t2_tse3d anatomical reference MR in Fusion). Note that the PET data has already been coregistered to the MR including reslicing.The data used in this case study was all from Siemens Inveon PET and for tracers labeled with C-11 - the performance of the model may vary for data from different hardware and/or with different tracers.