PAI is provided with a neural network architecture designed for multichannel segmentation. This architecture expects one or more 3D input series and generates segmentation results with one or more segments/VOIs.
The initial application of this architecture was for a case called Tumor Detection. A trained model BRATS Tumor Detection (Multichannel Segmentation) is available in the View and Segmentation tools when PAI is licensed. It is based on the MICCAI Brain Tumor Segmentation (BraTS) Challenge: http://braintumorsegmentation.org. BraTS utilizes multi-institutional pre-operative MRI scans and focuses on the segmentation of intrinsically heterogeneous (in appearance, shape, and histology) brain glioma tumors.
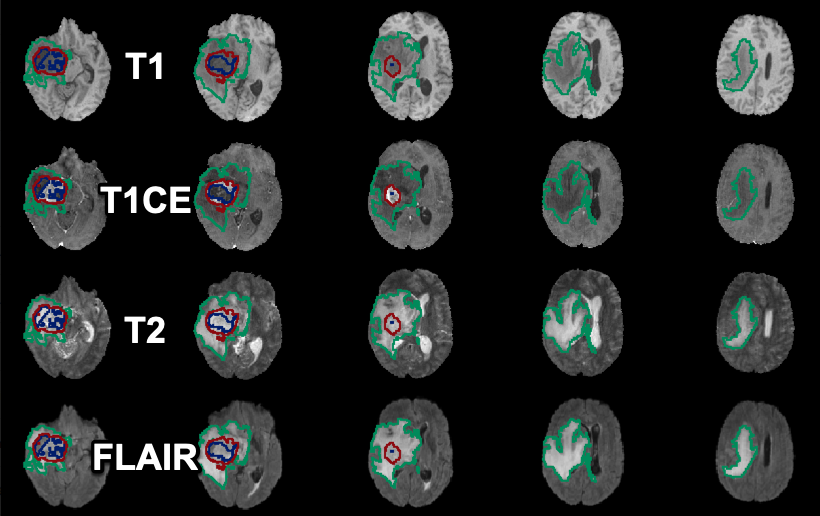
Training and testing of the Tumor Detection model in PAI was performed using the data from the 2020 BraTS Challenge containing 369 samples. Each sample consists of four MR images (native T1, post-Gd-contrast T1-weighted, T2 FLAIR, T2-weighted) and one image containing three reference segments as label numbers.
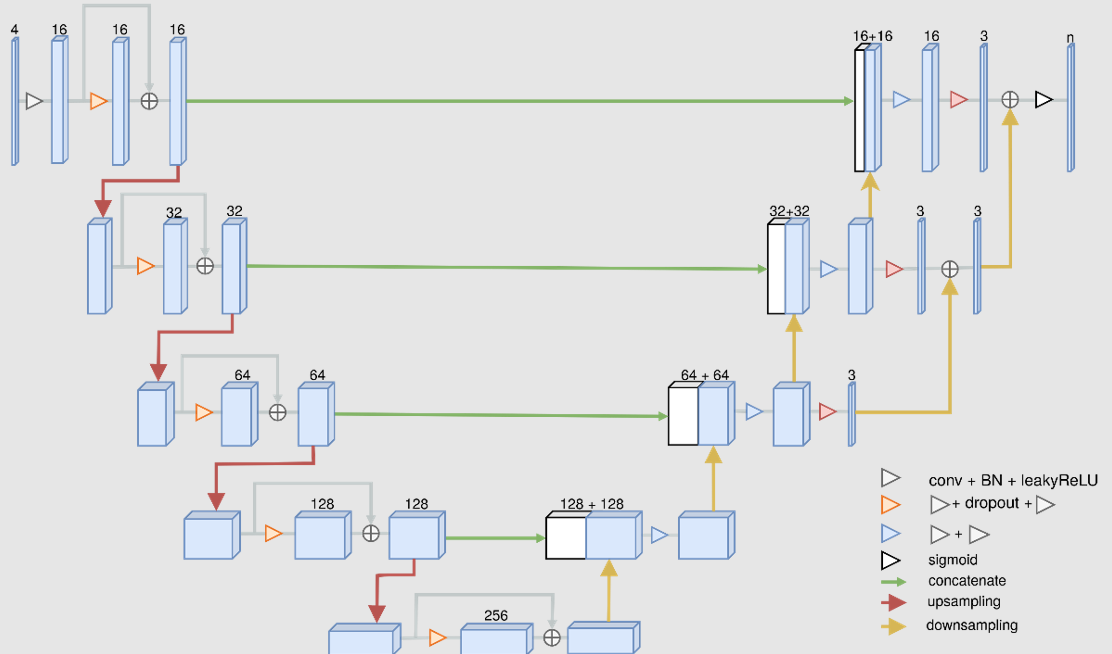
The Multichannel Segmentation architecture is a modified version of the convolutional neural network U-Net:
The output of the Tumor Detection model is a label image with three segments (label 1: non-enhancing tumor; label 2: peritumoral edema; label 4: Gd-enhancing tumor).
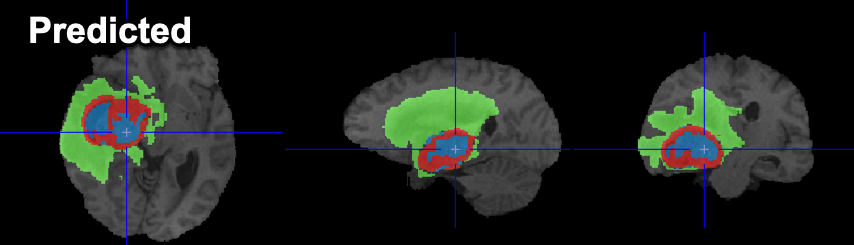
Example data to test the BRATS Tumor Segmentation model is available in our Demo database (Subject PAI1). This example is one of the subjects available in the data for the 2020 BraTS Challenge .
To try the model for yourself we recommend use of the Segment tool (PSEG). Select the T1 series as Input series. The T1CE, FLAIR & T2 series will be detected automatically according to the Association mechanism. The model selection must be BRATS tumor segmentation (Multichannel Segmentation). The model was trained with 3D data so no Split Slices/Frames is available or required. The data used in this case study was all from the BraTS Challenge and is skull-stripped 3D MR - the performance of the model may vary for data from different hardware and/or with different contrast/sequences/pre-processing.
BraTS Reference:
Bakas, Spyridon & Reyes Jan. (2019). Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. https://arxiv.org/pdf/1811.02629.pdf