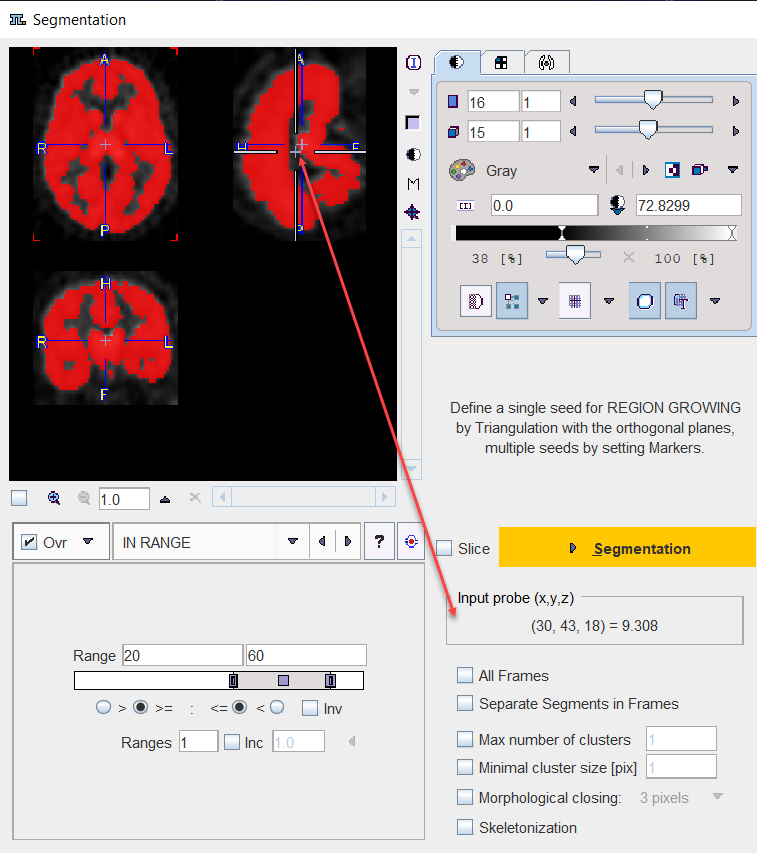
The Segmentation tool is a method for simple mask creation similar to the segmentation in the 3D tool. It opens the following dialog window.
Segmentation Page Layout
The Segmentation interface is organized as follows:
•The input images are shown in the upper left. An overly color is used to highlight the pixels which qualify for the current segmentation settings. This segmentation preview is only approximate.
•The segmentation method is configured in the lower left by a list selection. The parameters of the selected method are updated accordingly. There are several segmentation methods for finding the object contours.
•The actual segmentation is initiated with the Segmentation button. It produces a binary image in the upper left with a black background and the white object (depending on the segmentation type).
•The result is shown in the right image display port. Note that if the study is dynamic, an additional box All Frames appears. If it is checked, segmentation will be performed for all frames separately, otherwise with the frame which is currently shown.
•useful segmentation results can be saved as Template Atlas or outlined to VOIs and saved as .voi file.
Input Probe Value
Note the Input probe value which displays the image value at the position of the cursor, which is useful for finding appropriate Threshold setting.
Its value can be entered manually or using the Histogram panel for setting the threshold value.
Additional Segmentation Settings
•All Frames: this additional box appears only when the study to be segmented is dynamic. When enabled, the segmentation will be performed for all frames separately, otherwise with the frame which is currently shown.
•Segments in Separate frames: when enabled, allows organizing multi-segments results as a dynamic image: one segment per frame.
•Max number of clusters: when enabled it performs a cluster analysis and limits the number of clusters per segment according to the specified value.
•Minimal cluster size [pix]: when enabled it performs cluster analysis and includes in the segmentation results only cluster larger than the specified size.
•Morphological closing: when enabled it performs a morphological closing operation per segment according with the selected option: 3, 5 or 7 pixels.
•Skeletonization: The skeletonization algorithm allows extracting the "center-lines" of an object and uses them to efficiently represent the object. The skeletonization algorithm is complementary to the segmentation. This particular algorithm is described in a dedicated section.
Pixel Value Distribution
The Histogram in the lower right panel provides an overview of the pixel value distribution in the current image volume. It may indicate a peak usable for setting a background threshold. The current threshold value is indicated by the vertical line in the histogram.
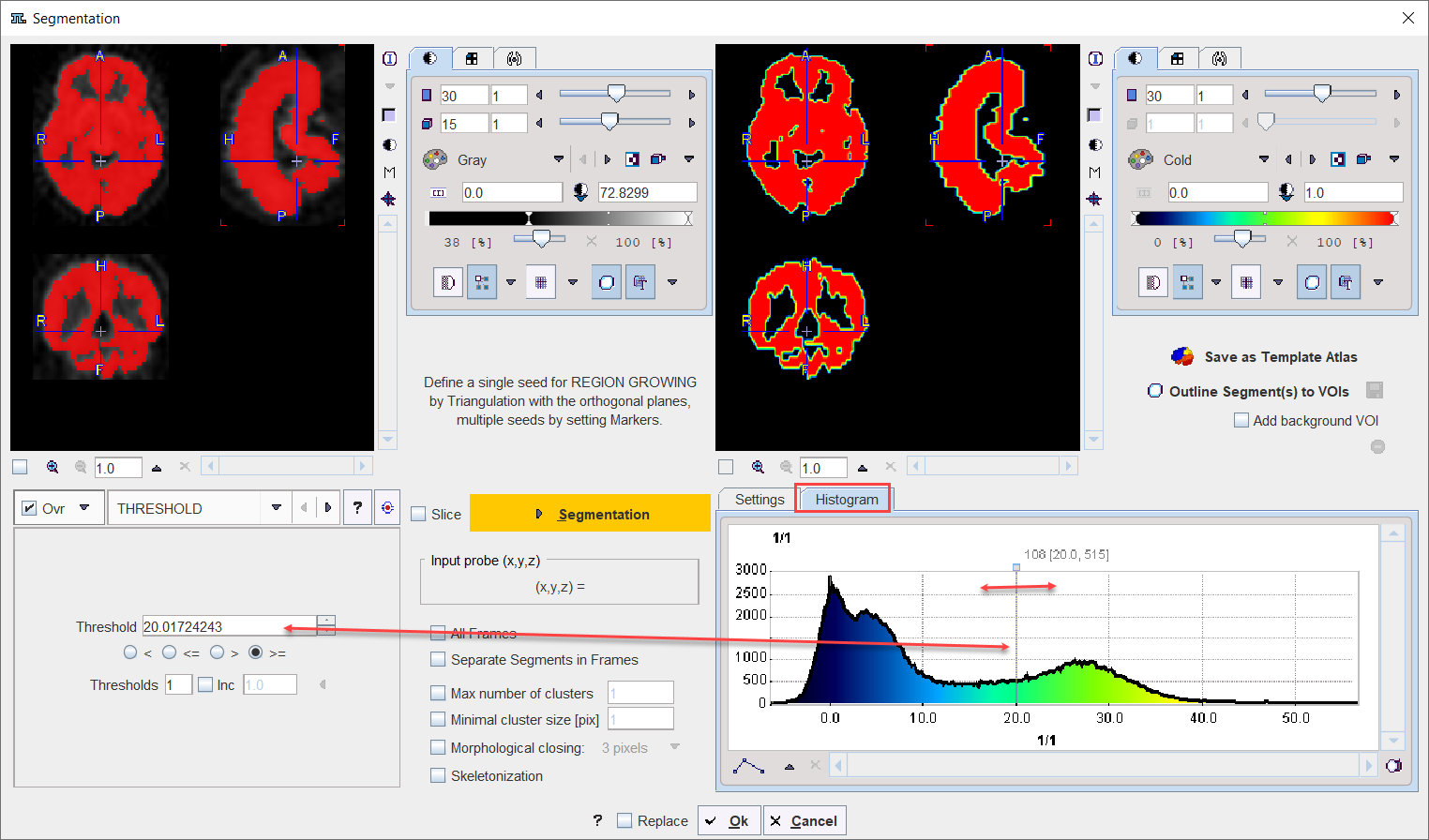
Note:
•When the study is dynamic the histogram content displays the pixel distribution of the currently selected frame. The Histogram panel content is updated when the frame is changed
•The histogram content is also updated when Restrict to VOI selection is enabled for the Segmentation VOI interface.
Segmentation Methods
The following segmentation methods are available.
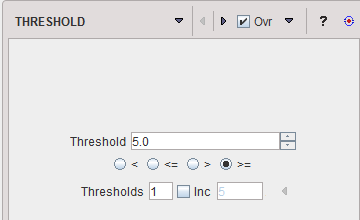
THRESHOLD |
The THRESHOLD segmentation is conceptually simple. All pixels with value above or below the threshold are included in the segment, depending on the condition settings.
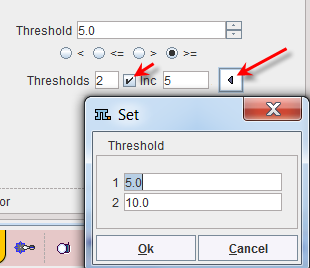
Sometimes it is helpful to segment at several threshold levels at once. This can easily be realized by setting the number in the Thresholds field accordingly, and entering the threshold values in the appearing Set dialog window.
Multiple threshold values can be incremented with a constant value enabling the Inc box and specifying the step in the dedicated text box. |
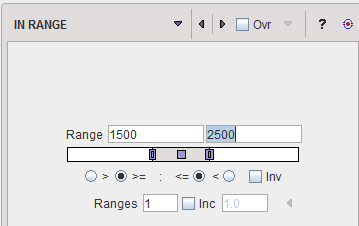
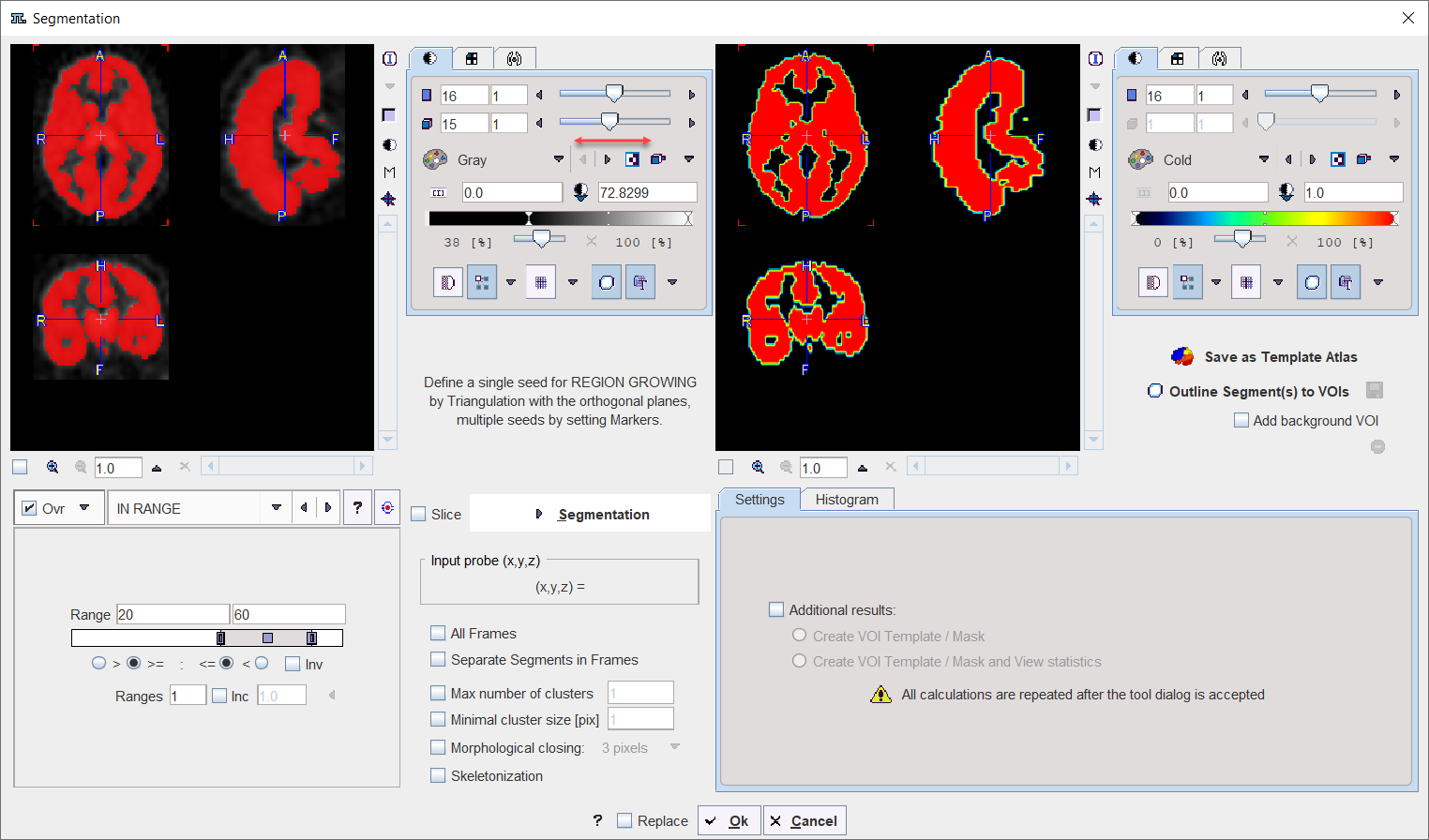
IN RANGE |
The IN RANGE segmentation is similar to the THRESHOLD method, except that two limits are defined. The boundary values are included or excluded, depending on the radio button settings. The complementary criterion is obtained by checking the Invert box.
Similar to the THRESHOLD segmentation, multiple Ranges can be defined and segmented at once. |
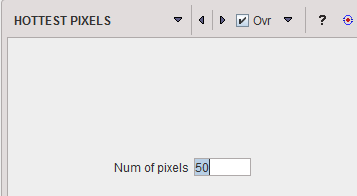
HOTTEST |
The HOTTEST PIXELS segmentation allows obtaining the (often disconnected) pixels with the highest values. The number of included pixels can be specified in the Num of pixels field.
|
REGION |
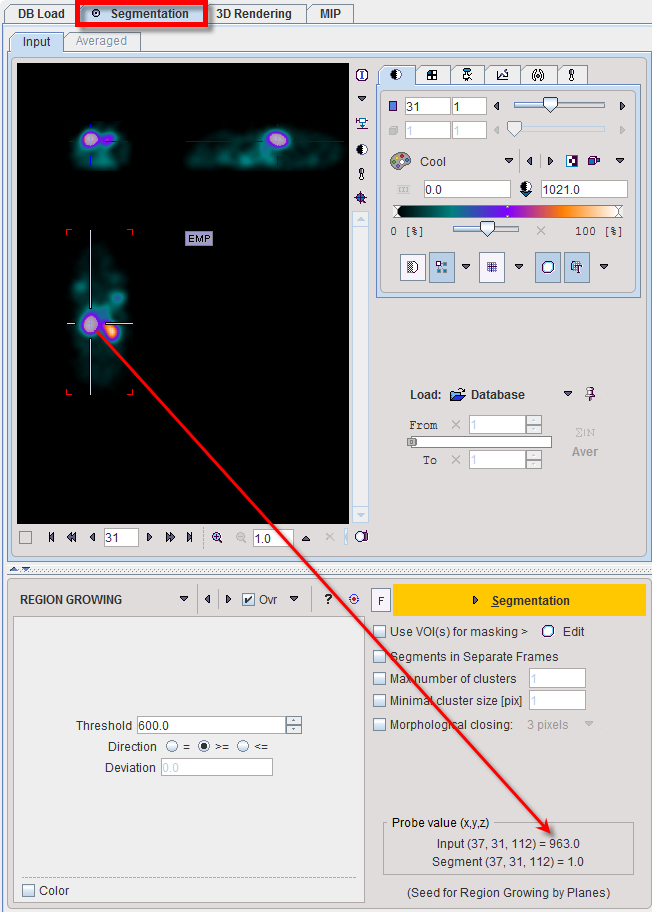
REGION GROWING is a method by which the user defines a starting point (seed) within the object of interest, and the algorithm tries to find all connected pixels which fulfill a certain criterion. It is required to triangulate the seed point with the orthogonal plane layout as illustrated below.
The inclusion criterion is formed by the Threshold value and the Direction specification: ▪= include all connected pixels with a value Threshold ± Deviation; ▪>= include all connected pixels above the defined Threshold value; ▪<= include all connected pixels below the defined Threshold value. Note the Probe value which displays the image value at the position of the cursor, which is useful for finding appropriate Threshold setting. |
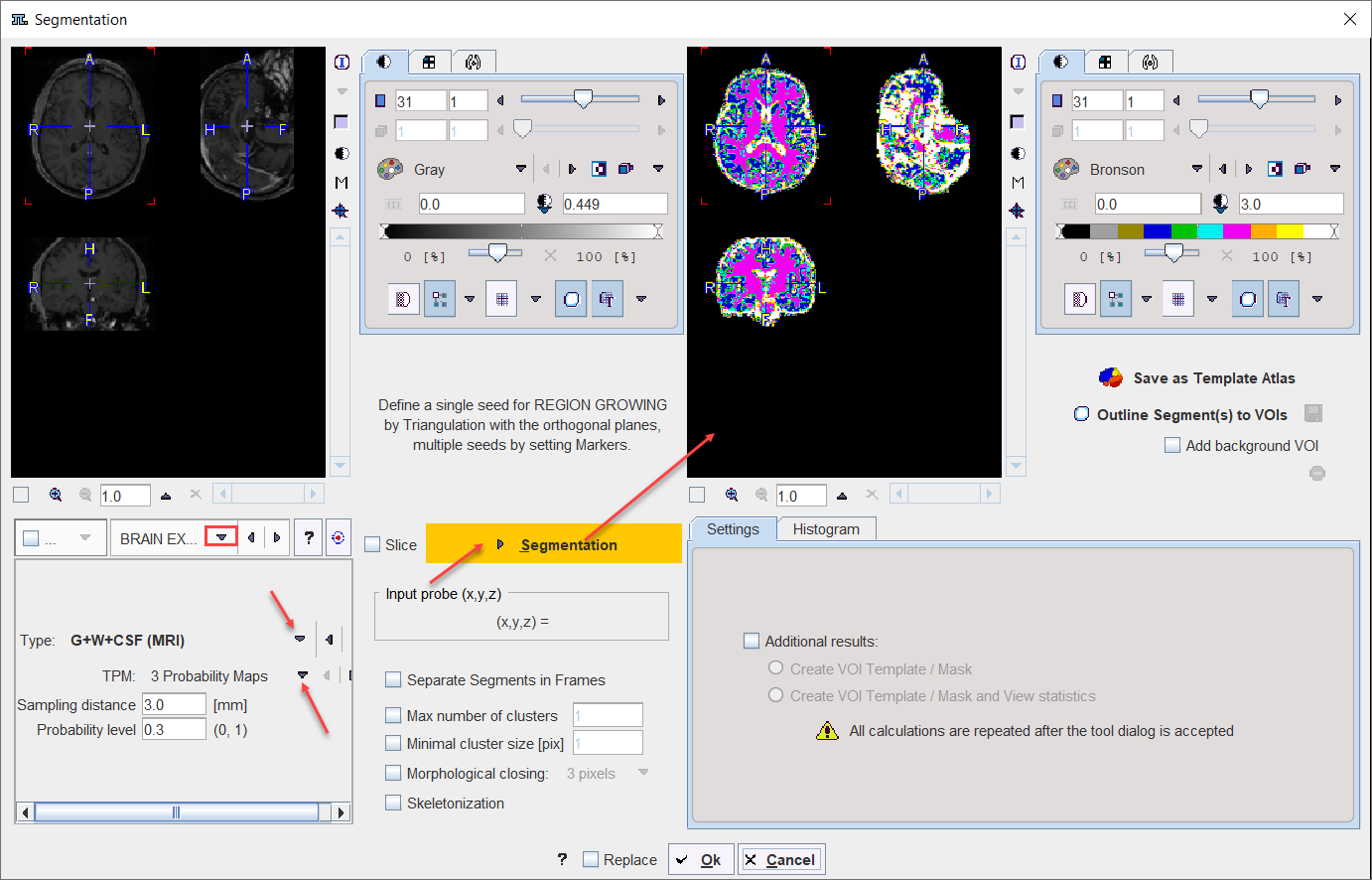
BRAIN EXTRACTION |
This segmentation can be used for 1.the extraction of the whole brain as a unique segment from a T1 weighted brain MRI. To this end the Brain (MRI) Type need to be selected. 2.the classification of the pixels in a T1-weighted brain MRI into Grey Matter, White Matter and CSF when the G+W+CSF is selected as Type. 3.the extraction of the whole brain as a unique segment based on the Mask of the selected Template, with parameter Sampling distance in mm and the Probability level for the map discretization.
Note: The methodology variants from point 1 and 2 above is an implementation of the segmentation in SPM8 (3 Probability Maps) and SPM12 (6 Probability Maps) respectively, with parameter Sampling distance in mm and the Probability level for the map discretization. |
K MEANS CLUSTERING |
K-Means is a popular method which partitions N observations into K clusters. The parameters are the Number of clusters (=K), the Number of iterations of the heuristic procedure, and an assumed Percentage of background pixels.
The procedure can be applied to static and dynamic data. In the latter case, pixel with similar signal shapes will be clustered using the time-weighted Euclidean distance as the measurement of dissimilarity (or distance) between TACs. Note that the All frames box has to be enabled, because otherwise each frame of the dynamic series will be clustered separately. The procedure for clustering based on the signal shape proceeds as follows: 1.Background pixels are removed by calculating the signal energy of the pixel-wise TACs (sum of squared TAC values), and considering only pixels above a specified percentile. 2.K non-background pixels are randomly selected as initial cluster centroids . 3.Each non-background pixel is assigned to the centroid with minimal distance between the TACs, thus forming K initial clusters. 4.For each cluster a new centroid TAC is calculated as the average TAC of all pixels in the cluster. 5.An iterative process is started which repeats the following two steps: (1) Each pixel TAC is compared with all centroid TACs and assigned to the cluster with minimal distance. (2) All centroid TACs are recalculated to reflect the updated cluster population. The iterations are repeated until no pixels are re-assigned to a different cluster, or a maximal number of iterations is exhausted. |
SCATTER in VOI |
This particular method for localizing pixels from 2D scatter plots is described in a separate section. |
ACTIVE MODELS |
Please refer to the dedicated Active Models description for details. |
HOTTEST CONNECTED VOXELS |
Is a segmentation method used to objectively outlined brain anatomical structure like the locus coeruleus/subcoeruleus complex in T1 weighted MRI images. Another potential application is the extraction of the image derive input function. Please refer to the dedicated Hottest Connected pixels topic for details. |
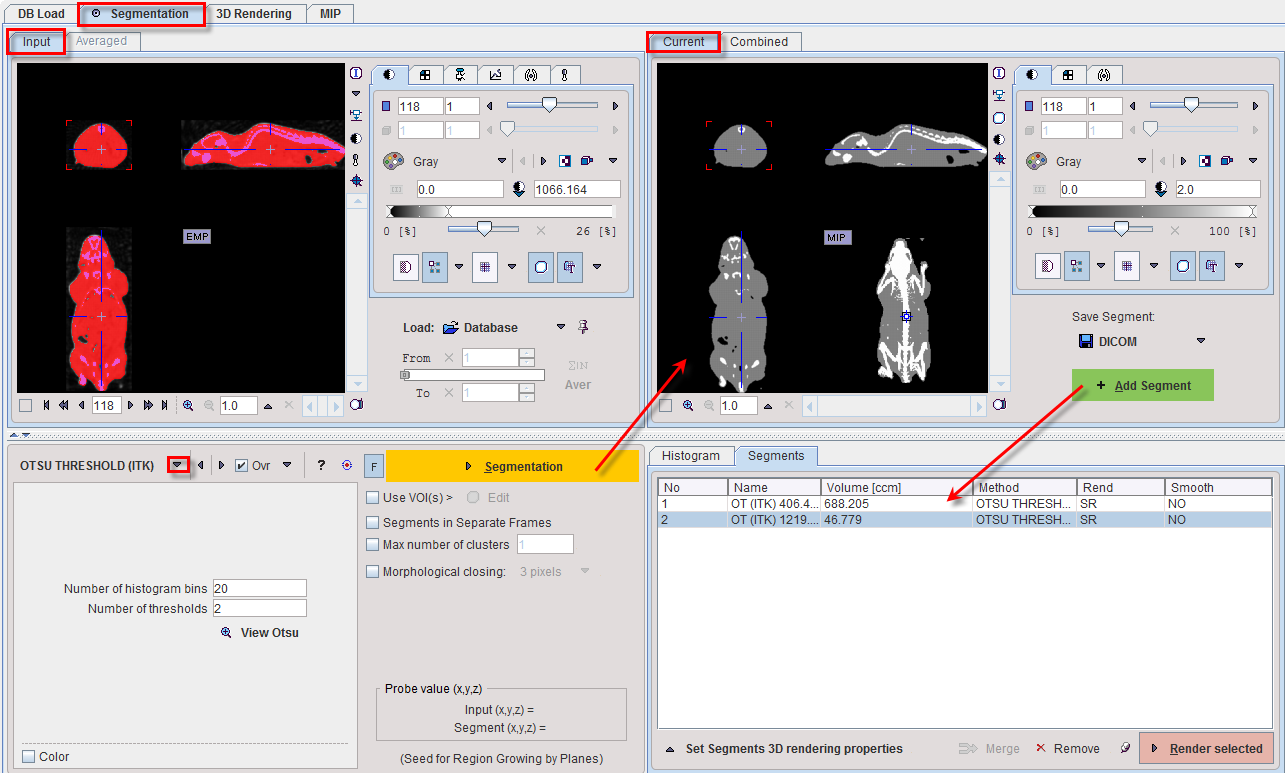
OTSU THRESHOLD |
The OTSU THRESHOLD method is based on the analysis of the pixel histogram and is described by The ITK Software Guide as follows: "Another criterion for classifying pixels is to minimize the error of misclassification. The goal is to find a threshold that classifies the image into two clusters such that we minimize the area under the histogram for one cluster that lies on the other cluster’s side of the threshold. This is equivalent to minimizing the within class variance or equivalently maximizing the between class variance." The Number of histogram bins defines the resolution of the histogram shape, whereas the Number of thresholds defines the number of distinct clusters. With the mouse CT, Otsu with the settings below finds appropriate thresholds for the skeleton object and the soft tissue, as illustrated below. Note that the threshold values can be inspected only after the actual segmentation by the View Otsu button.
|
MACHINE LEARNING |
If the Pmod Artificial Intelligence (PAI) module has been licensed, the MACHINE LEARNING segmentation options will be available. Please refer to the dedicated PMOD Artificial Intelligence Framework (PAI) user guide for detailed information. |
Segmentation Results Saving
The segmentation result can be used in different ways:
▪Save as Template Atlas directly saves the segmented series in an atlas format in the directory Pmod4.4/resources/templates/voitemplates. Subsequently, it can be used for statistics using the Atlas tab of the Templates pane in the VOI definition function.
▪The Outline Segment(s) to VOIs button allows creating VOIs for each generated segment. Optionally, the background can be added as a VOI by enabling the Add background VOI box. The outlined VOIs can be saved as files and later used for statistics by the Save icon next to the Outline button.
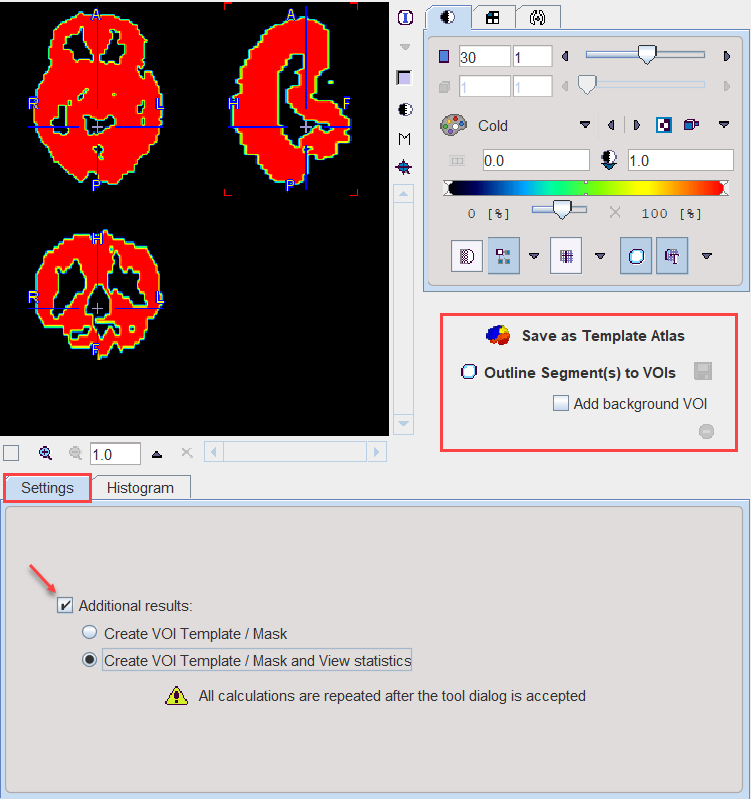
With the Ok button the segmentation is performed again and the resulting segment returned as an image series which is binary for a simple segmentation and an object map in the case of a clustering method. The following Additional results can be returned.
1.Create VOI Template/Mask: With this setting, the segments are also returned as a VOI template which can be activated by the Mask tab of the VOI Template function.
2.Create VOI Template/Mask and View Statistics: As 1), but the statistics on the current image are additionally calculated.