Rapid Model
The original resolution of the samples was 5.5 and 2.8 um and for training all were resampled to 50 um. No additional cropping was applied and the image values were normalized using the z-score method. Training was performed as follows:
notebook computer with 8 core 2.3 GHz Intel i7 and 32 GB RAM using only CPU
•70 samples (48 training, 12 validation, 10 testing)
•batch size 12
•learning rate 0.0005
•300 epochs
Training took 151 seconds. The training loss value was 0.0368 and the validation set loss value was 0.0388. Testing was performed on the 10 additional samples using the Evaluate Model functionality with a loss value of 0.1037.
The model was tested on the second femur dataset with reduced pixel size of 0.0112 mm, and on the eighth tibia dataset with native 0.005 mm pixel size. Both datasets were loaded as 3D volumes and prediction was run with the Split Slices option. The trabecular region was correctly segmented in < 20 seconds on a notebook similar to that used for training.

Due to training with resampling to 50 um the resulting segments displayed clear staircase artefacts. There was some overlap with cortical bone. A segment smoothing approach was investigated to remove the staircase artefacts. The segment was transferred from the Segment tool to View and smoothed with a Gaussian filter at 0.05 x 0.05 x 0.05 mm FWHM. Automatic isocontouring at 50% (Max-min) threshold was used to create a new VOI.
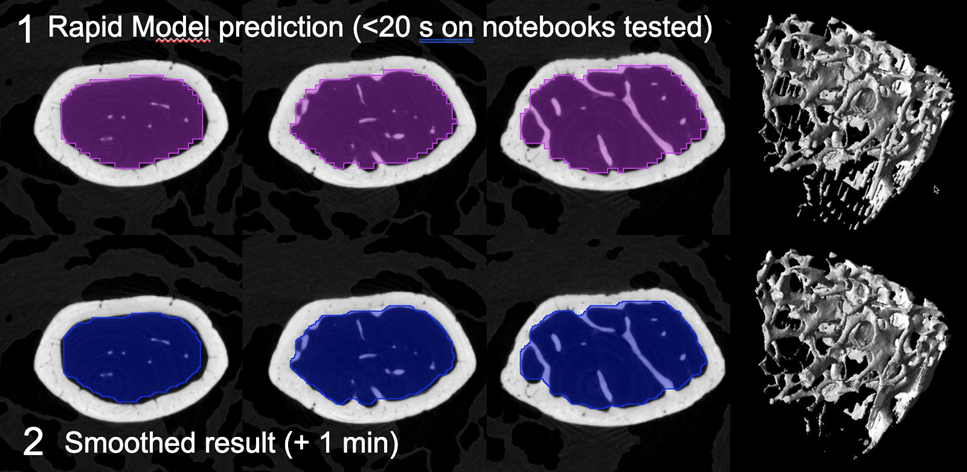
Smoothing visually improved the VOI but some overlap with cortical bone remained.
On tibia segmentation also took < 20 seconds on the notebook tested. Similar staircase artefacts and some overlap with cortical bone were present.
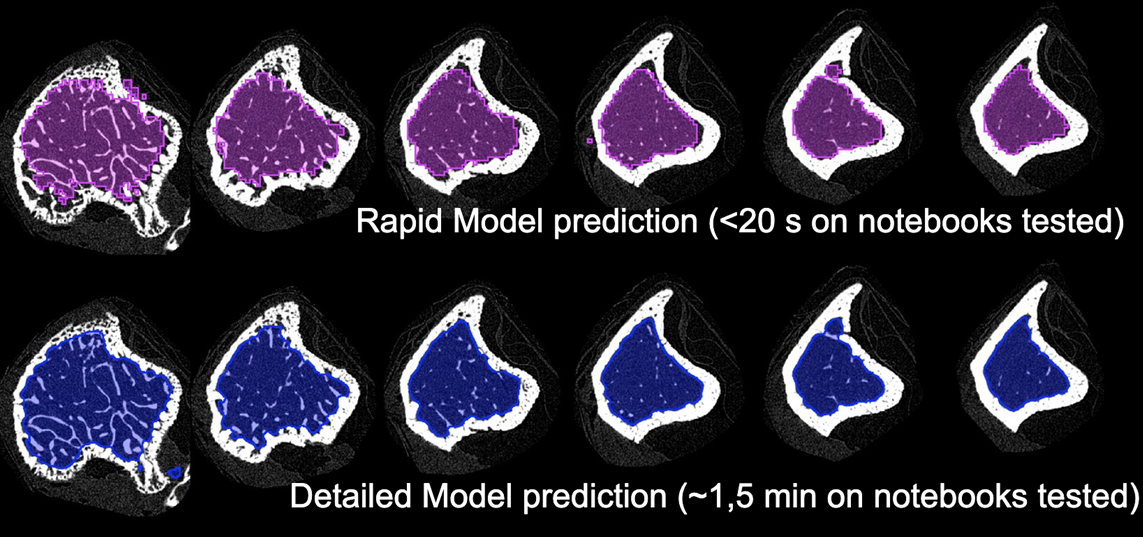
Detailed Model
The original resolution of the samples used was 5 and 5.5 um and for training all samples were resampled to 10 um. No additional cropping was applied and the image values were normalized using the z-score method. Training was performed as follows:
•AWS p2.8xlarge, 96 GB GPU, 488 GB RAM
•3509 samples (2486 training, 622 validation, 401 testing)
•batch size 60
•learning rate 0.0005
•10 epochs
Training took 600 seconds. The training loss value was 0.0546 and the validation set loss value was 0.0545. Testing was performed on all slices of the eighth tibia dataset using the Evaluate Model functionality with a loss value of 0.0580.
The trained model was tested on the same femur and tibia datasets as the rapid model. Both datasets were loaded as 3D volumes and prediction was run with the Split Slices option. Prediction on the tibia in native pixel size took approximately 90 seconds on the notebook tested. The contour closely followed the endosteum (inner tibial surface) with some artefacts present close to the growth plate. On femur several artefacts were present that resulted in inner holes in the trabecular VOI. Manual editing was used to close the holes and allow morphometric analysis.
Morphometric analysis
The segments generated by both models and smoothed versions of the rapid model segments were used for further trabecular bone analysis. A single threshold of 2000 Hounsfield units (HU) was used to segment trabecular bone voxels within the model segments. The parameters BV/TV (bone / tissue volume), trabecular thickness, trabecular number, trabecular separation, and fractal dimension were calculated using the volume and surface area statistics according to the plate model.
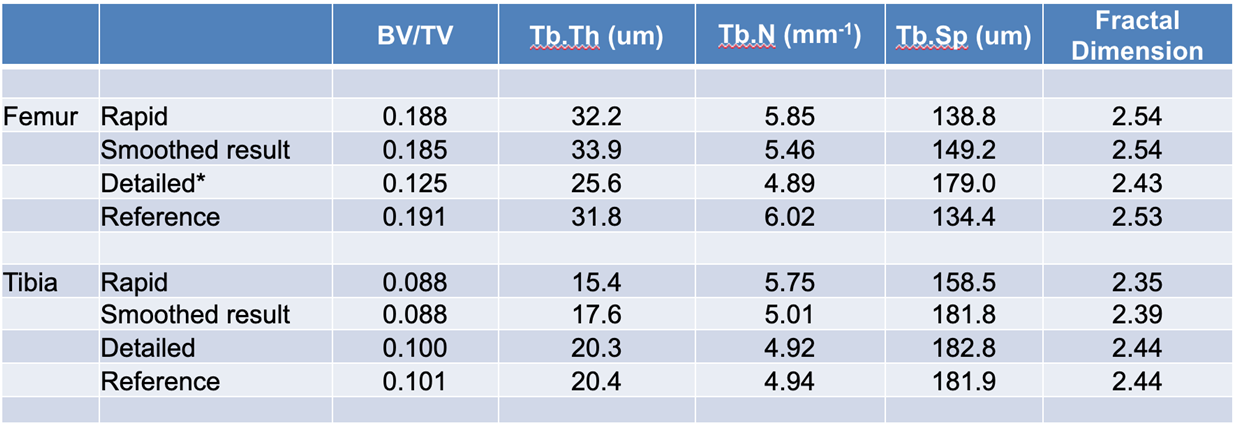
Morphological analysis revealed that the rapid model yielded comparable results to the reference segments for the femur tested, but resulted in marked deviation from the reference segments for the tibia tested. Smoothing of the segments generated by the rapid model did not universally improve the result. The detailed model resulted in near replication of the reference results for tibia, but did not produce good results for the femur. We believe that this was likely a result of overtraining the model towards tibia (seven tibia vs. one femur in training set).
Use of either model for study-level trabecular analysis appears feasible. Erosion of the tissue volume segment resulting from PAI by several voxels in 2D mode would likely be beneficial to further ensure that cortical bone is excluded.
Example data to test the Mouse bone trabecular model is available in our Demo database (Subject PAI6). The data used for the case study was kindly provided by Dr. Phil Salmon from Bruker microCT.
To try the model for yourself we recommend use of the Segment tool (PSEG) or segmentation interface in View. Either the low res or full res series may be used for testing. The low res series has been downsampled but includes more slices in the 3D volume. The model selection must be Trabecular ML (uNET Segmentation). The model was trained with 2D data so Split Slices is required. No further cropping of the demo data is required. The data used in this case study was from Bruker microCT - the performance of the model may vary for data from different hardware and/or with different reconstruction/pre-processing.