A common application in bone research is analysis of bone remodeling in the trabecular of distal femur and proximal tibia in mice. MicroCT imaging of ex vivo femur and tibia is used to generate 3D datasets with pixel size in the range of 5 um. Analysis of trabecular bone is then performed in the metaphysis and requires an initial segmentation separating trabecular bone from cortical bone and the growth plate. Simple cropping of the 3D dataset is usually sufficient to avoid the growth plate and to define the division between metaphysis and diaphysis. Some researchers additionally distinguish between primary and secondary spongiosa.
Definition of the trabecular region to be analysed, including as much material as possible but without including cortical bone, can be laborious and even automated solutions need tedious clean up. AI-based segmentation could offer a rapid alternative and provide the starting “tissue volume” for further trabecular analysis.
We used two distal femur and eight proximal tibia microCT datasets to train a uNet architecture using two approaches. In a strategy to increase the amount of data available for training, we treated each 2D axial slice of the femur/tibia as a separate sample. One femur and all eight tibia datasets were provided with reference segments created in the Bruker microCT software CTan.
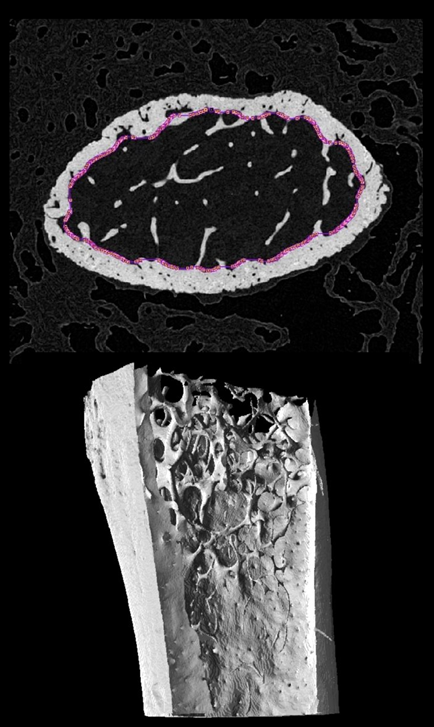
Example data to test the Mouse bone trabecular model is available in our Demo database (Subject PAI6). The data used for the case study was kindly provided by Dr. Phil Salmon from Bruker microCT.
To try the model for yourself we recommend use of the Segment tool (PSEG) or segmentation interface in View. Either the low res or full res series may be used for testing. The low res series has been downsampled but includes more slices in the 3D volume. The model selection must be Trabecular ML (uNET Segmentation). The model was trained with 2D data so Split Slices is required. No further cropping of the demo data is required. The data used in this case study was from Bruker microCT - the performance of the model may vary for data from different hardware and/or with different reconstruction/pre-processing.