A new Learning Set was prepared, containing the 369 available samples, configured for a segmentation task and using the Multichannel Segmentation architecture.
An R Workspace containing all samples was exported and used for training on commercial cloud computing infrastructure. Training was performed with the settings:
•AWS p3.8xlarge, 4 V100 GPU, 244 GB memory
•369 samples (295 training, 74 validation)
•Crop to associated VOI
•500 epochs
•batch size of 4
•Learning rate 0.0005
The final training set loss value was 0.2103 (1 - Dice coefficent) and the validation set loss value was 0.3531. Training took 810 minutes.
The weights and manifest resulting from the training were retrieved and used to add the training model to the /resources/pai folder for deployment on local installations.
An example of the results on one subject are illustrated below:
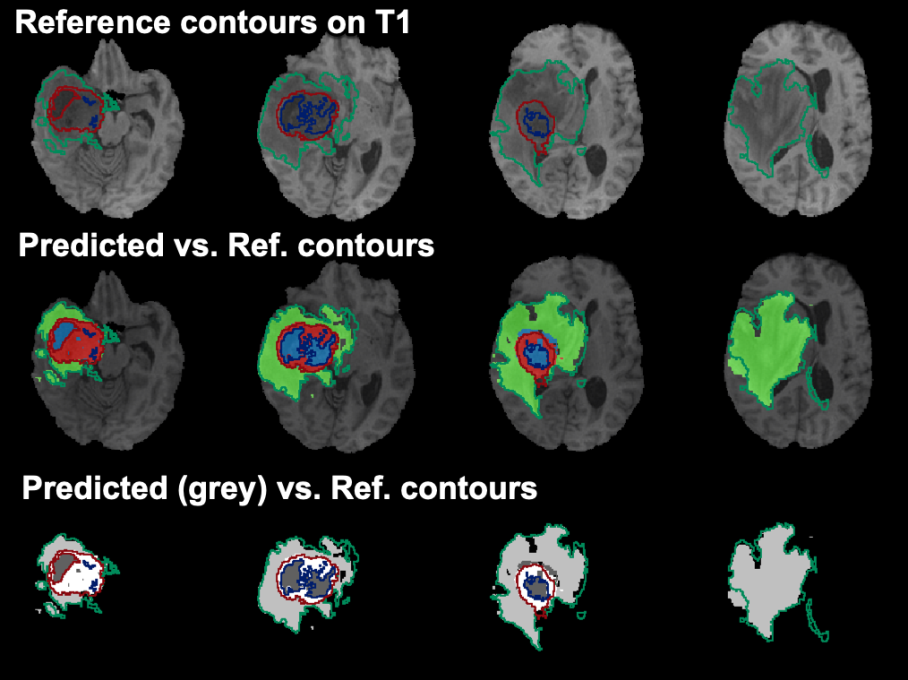
Example data to test the BRATS Tumor Segmentation model is available in our Demo database (Subject PAI1). This example is one of the subjects available in the data for the 2020 BraTS Challenge .
To try the model for yourself we recommend use of the Segment tool (PSEG). Select the T1 series as Input series. The T1CE, FLAIR & T2 series will be detected automatically according to the Association mechanism. The model selection must be BRATS tumor segmentation (Multichannel Segmentation). The model was trained with 3D data so no Split Slices/Frames is available or required. The data used in this case study was all from the BraTS Challenge and is skull-stripped 3D MR - the performance of the model may vary for data from different hardware and/or with different contrast/sequences/pre-processing.