k-means clustering of dynamic data is working completely data-driven without any prior knowledge apart from the number of clusters. The SUPERVISED clustering approach is complementary. Here the user specifies a set of TACs which are characteristic for the kinetics of different tissues. The algorithm then decomposes the data into clusters with kinetics similar to the prescribed TACs.
This method was introduced by Turkheimer et al. [8] for determination of gray matter tissue without specific binding as a reference in [11C]PK11195 brain studies. It requires preparation of time-activity curves which are representative for different tissues in the brain. For [11C]PK11195 the authors found that the method worked best after normalizing the data frame-wise by subtracting from each pixel value the frame average and dividing by the frame standard deviation. Note that this pre-processing is not part of the CLUSTER ANALYSIS (SUPERVISED) method. Rather, the images have to be normalized beforehand using the z-score external tool. As a result of this normalization, the image characteristics change remarkably.
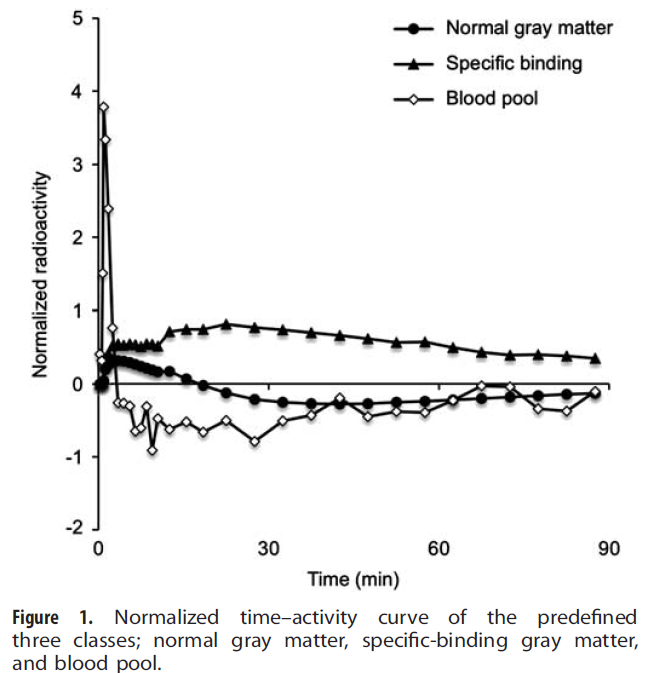
The implementation in PSEG is close to the supervised clustering variant presented by Ikoma et al. [9] for use with [11C]PIB. Particularly, the weight ratio (eq. 3 in [9]) is used to assign each pixel to one of the classes, or to background. The illustration below shows the classification TACs used for normalized [11C]PIB data [9].