Starting from the operational equation of the blood-based Logan plot, Ichise et al. derived three multi-linear reference tissue model variants MRTM0, MRTM and MRTM2 [1]. They all assume an initial equilibration time t* from which on the derived multi-linear relation holds. However, if kinetics in the target tissue can be described by a 1-tissue compartment model (an assumption required for the SRTM), all data can be used for the fitting (t*=0). Otherwise an adequate t* value has to be determined.
Assuming the presence of receptor-devoid reference region TAC CT'(t), the target tissue TAC CT(t) is plotted as a function of the transformed tissue TACs as illustrated below. For the calculation of BPND it is assumed that the non-displaceable distribution volumes in the tissue and reference regions are identical.
When applied to noisy data such as single-pixel TACs in parametric mapping, the MRTM method still suffers from a high variability. Assuming a known value of the reference tissue clearance rate k2' the MRTM operational equation can be reformulated as the MRTM2 operational equation:
![]()
with only two regression coefficients VT/(VT'b) and 1/b for T > t*. The multi-linear relationship above can be fitted using multi-linear regression, yielding three regression coefficients. The binding potential is then calculated from the ratio of the two regression coefficients as
![]()
For receptor ligands with 1-tissue kinetics such as [11C]DASB the multi-linear equation is correct from t*=0, and the clearance rate constant from the tissue to plasma k2 is equal to the negative value of the second regression coefficient, -(1/b). Furthermore, R1 = K1/K'1, the relative radioligand delivery, equals the first regression coefficient divided by k2'.
For pixelwise applications the same two-step approach applied in the SRTM2 model is applied:
1.Calculate in the model preprocessing step the clearance rate k2' of the reference TAC by the MRTM method with VOI data which has a limited level of noise.
2.Fix k2': Use the estimated k2' value for the pixelwise MRTM calculations, reducing the number of fitted parameters from 3 to 2.
Alternatively, k2' can be determined for externally (for instance in PKIN), manually entered and fixed.
Acquisition and Data Requirements
Image Data |
A dynamic data set acquired long enough that the equilibrium relation is approximately fulfilled. |
Target tissue |
Optional: TAC from a receptor-rich region (such as basal ganglia for D2 receptors). For visualization of model fitting and optionally for fitting k2' and t* |
Reference tissue |
Mandatory: TAC from a receptor-devoid region (such as cerebellum or frontal cortex for D2 receptors). Note: specification of an appropriate reference TAC is crucial for the result! |
VOI |
Optional: VOI definition excluding the reference tissue which can be used for getting an estimate of k2'. |
Model Preprocessing
t* |
The least squares estimation should be restricted to a range after an equilibration time. t* marks the beginning of the range used in the multi-linear regression analysis. It can be fitted based on the Max. Err. criterion if a Target tissue is specified. |
Max. Err. |
The maximal relative error allowed if t* is fitted. |
R1 Cutoff |
Alternative method for masking based on the R1 estimate. |
k2' for mapping |
k2 of the reference tissue. It can be specified in four different ways, as explained in section Specification of k2'. |
Percent masked pixels |
Exclude the specified percentage of pixels based on histogram analysis of integrated signal energy. Not applied in the presence of a defined mask. |
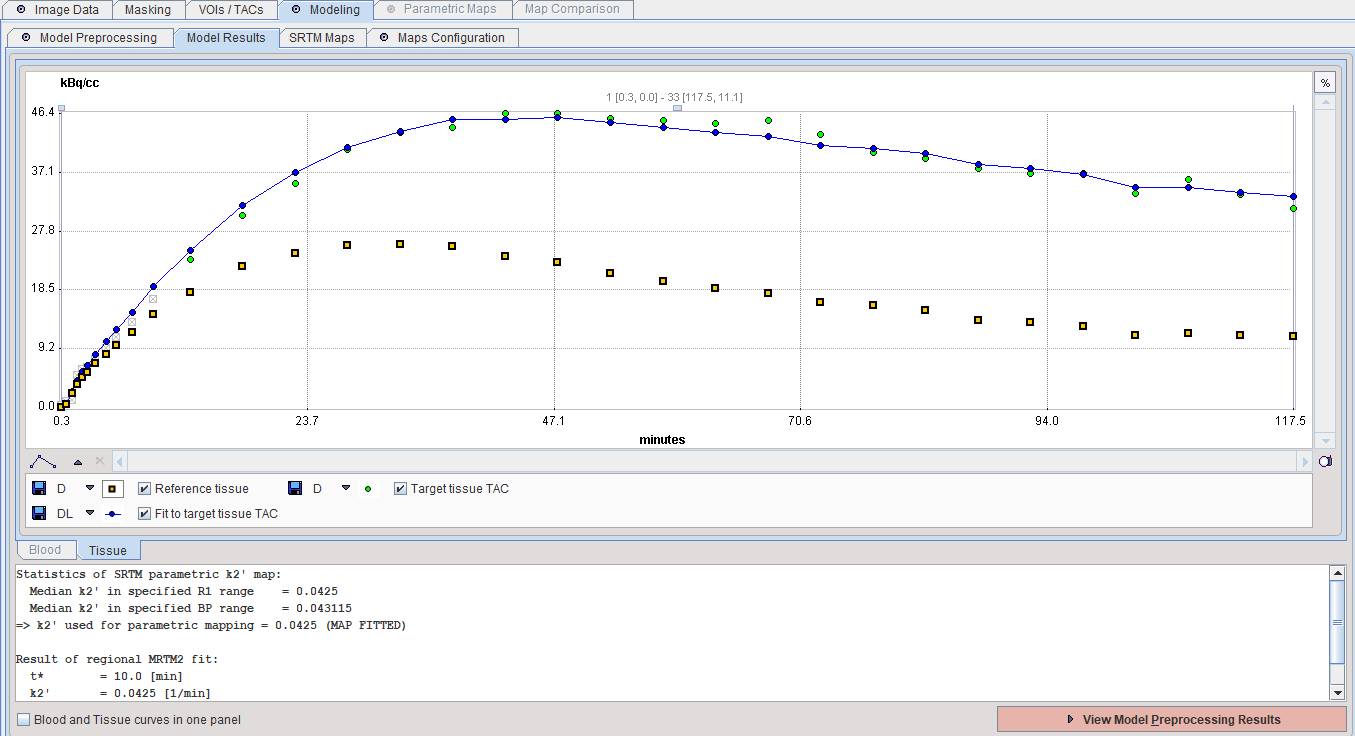
The result of a model fit during Model Preprocessing is shown in the Model Results panel for inspection. The initial points which are not taken into account (before the t* time) are indicated in grey. If no Target tissue is specified, the panel remains empty.
Model Configuration
![]()
BPnd |
Binding potential BPnd = k3/k4 . |
R1 |
R1 = K1/K1' relative ligand delivery (for 1-tissue kinetics). The calculated image often looks similar to a perfusion image and can sometimes used for matching purposes. It is recommended to restrict the range of fitted values. |
k2 |
Clearance rate in the pixel (for 1-tissue kinetics). |
Reference
1.Ichise M, Liow JS, Lu JQ, Takano A, Model K, Toyama H, Suhara T, Suzuki K, Innis RB, Carson RE: Linearized reference tissue parametric imaging methods: application to [11C]DASB positron emission tomography studies of the serotonin transporter in human brain. J Cereb Blood Flow Metab 2003, 23(9):1096-1112. DOI