The New button in the Macro line opens the editor illustrated below for creating new macros.
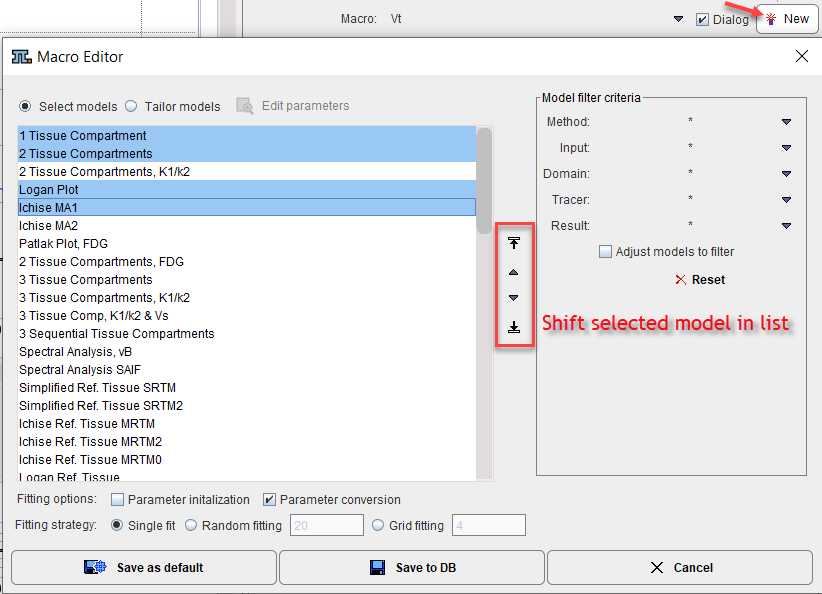
Models in the Macro
Use the Select models radio button to assemble the models for a macro. The available models are listed to the left. This list can be reduced by the filtering options to the right explained in the Model Filtering section. The list can be reordered by selecting an entry and shifting it with the arrow buttons indicated above. Once the model order is proper, select the models to form the macro in the list. Note that the models are fitted sequentially in list order, and the results are stored chronologically in the model history. Consequently, at the end of macro execution, the result of the last model will be shown.
Initial Parameters
To tailor the macro models to a certain data analysis task the initial parameter can be edited. Activate Tailor models to reduce the list to the selected models. Now a model can be selected, and its initial parameters as well as the restrictions tailored with the Edit parameters functionality.
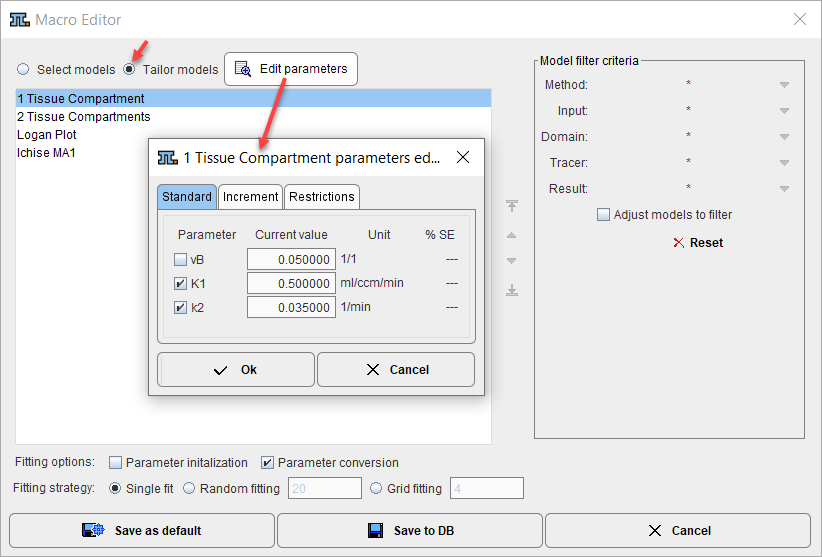
Fitting Options
Macros store also the configuration of the fitting options available in the lower section. During macro execution they will be applied for all models as applicable. Valuable uses of Parameter conversion include propagation of a common vB, k2' or t* as detailed here.
Macro Saving
Once the models are configured and the fitting options specified, the macro can be saved in two ways:
1.Save as default: This will save the macro in the resources/pkin/macros directory
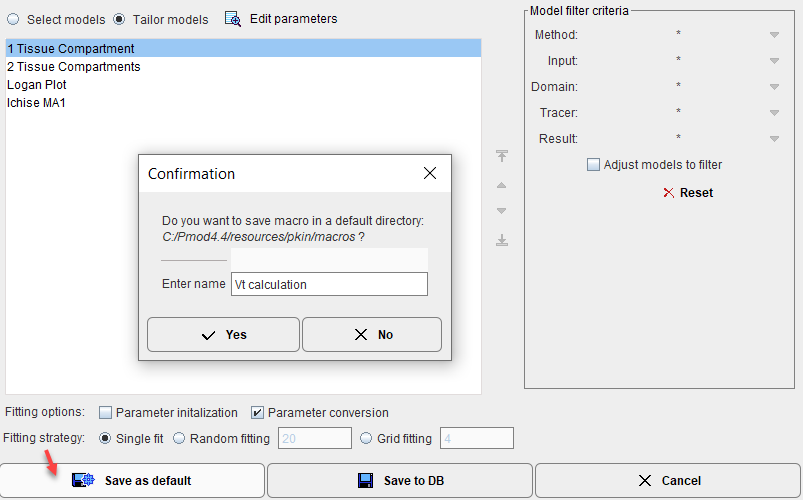
and as a result the macro will immediately appear in the Macro list
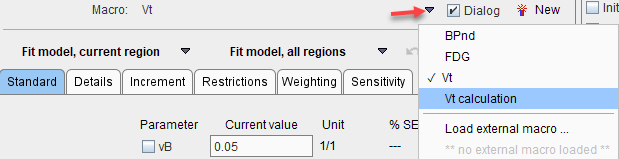
Note: For cleanup of the list please delete unnecessary .kmMacro files from the resources/pkin/macros directory in the PMOD installation.
2.Save to DB: This allows saving the macro in a PMOD database, or via Save to File System anywhere on a disk.