In certain circumstances, the matching algorithm alone may not be sufficient for a successful result. If this happens, the user can define masks for the reference and /or input image after loading. Masking will exclude pixels outside selected areas from calculations.
There are three options available for the mask definition:
1.Using the ...Create mask![]() icon (display definition).
icon (display definition).
2.Loading an existing mask using the ![]() icon (display definition).
icon (display definition).
3.Defining the mask in the Basic parameters tab available for the automatic matching algorithms.
The procedures are described below.
1. Create Mask (display definition).
To create a mask activate the ...Create mask ![]() icon as shown below.
icon as shown below.
Initially the masking interface appears with the loaded image in the left display area.
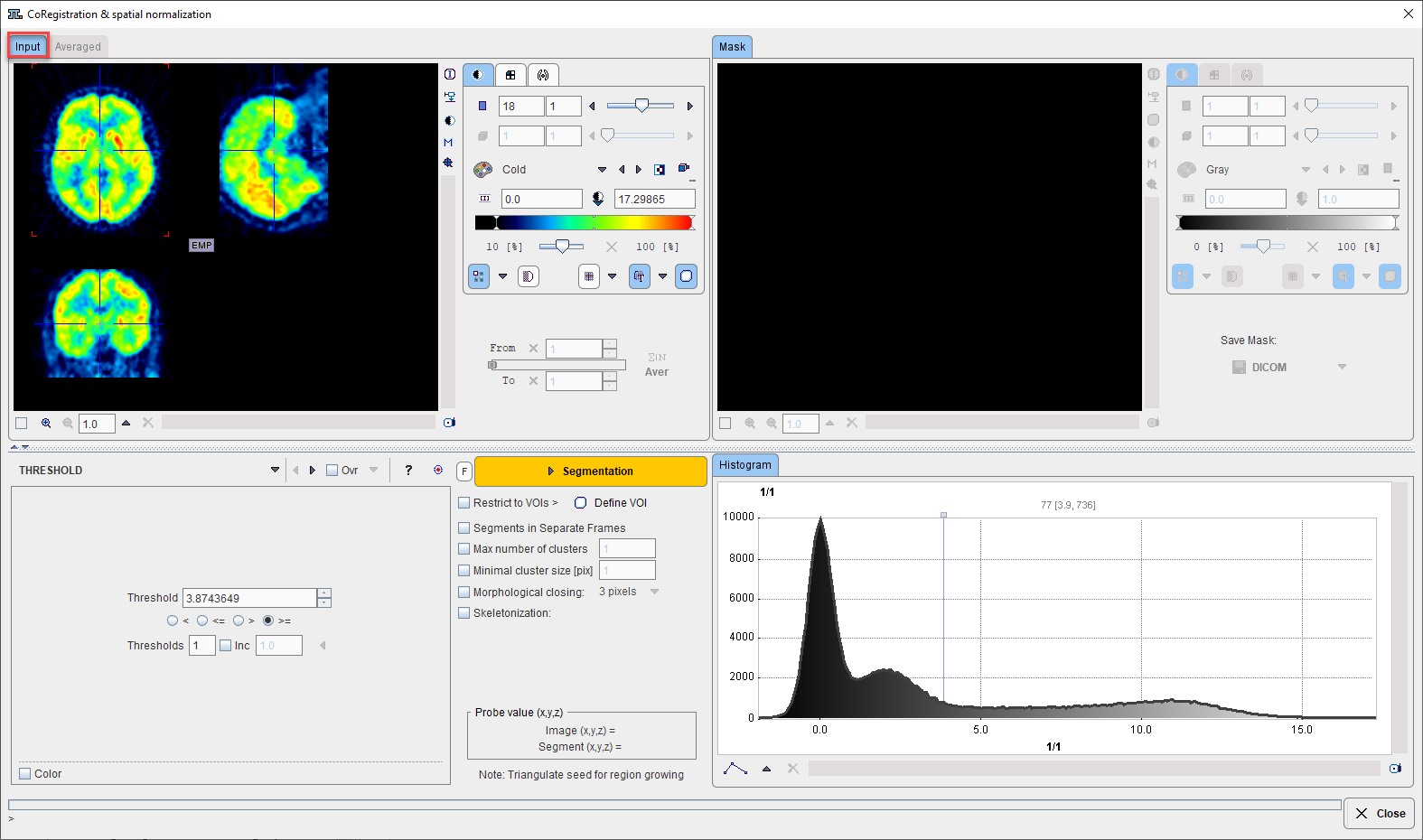
When working with dynamic series, it is recommended to average within an appropriate range in order to obtain an appropriate data set for masking. The range can be specified by the From and To numbers or using the slider handles. When the Average button is activated, the average uptake in the specified frame range is calculated and the result image is shown on the Averaged sub-pane.
Segmentation for Creating a Mask
The next step consists of generating segments which represent tissues of interest. Segmentation can be performed on Input series but also on the Averaged images, depending on which tab is selected. The Histogram of the pixel values is updated according to the selected images.
It is recommended to change the color table to Gray, and to enable the overlay Ovr box. Then, select one of the segmentation methods (described below) to specify an inclusion criterion. The pixels which satisfy the criterion are colored in red in the image overlay. Note that overlay updating might be slow when changing a segmentation parameter, depending on the segmentation method. Segmentation performs the actual segmentation and shows the result in the Mask tab to the right. While standard segmentations create binary images with 0 (background) and 1 (segment) pixel values, there are clustering approaches which generate multiple segments in a single calculation. These segments are distinguished by increasing integer pixel values. Each Segmentation activation overrides the previous contents in Mask pane.
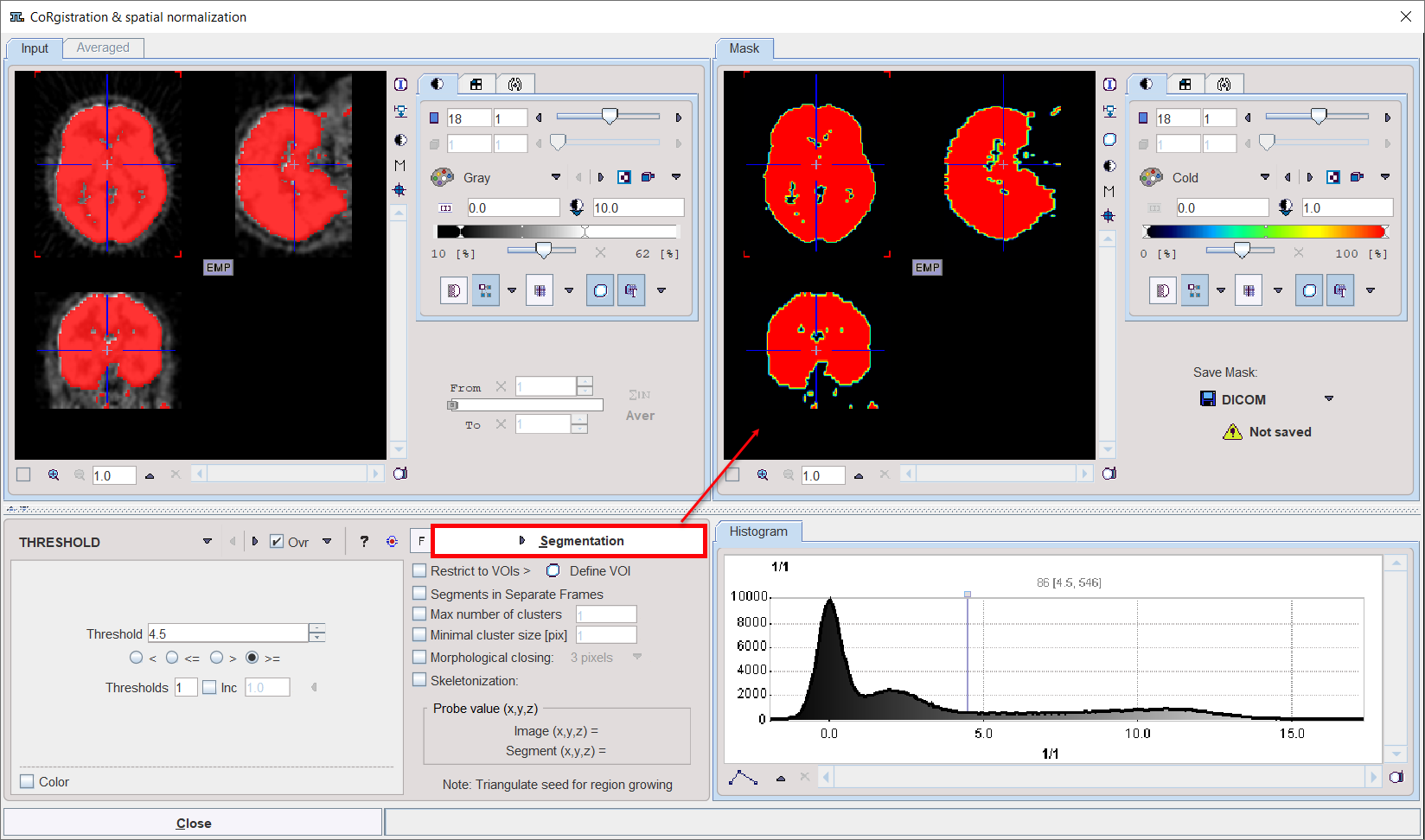
In certain circumstances, the segmentation methods alone may not be sufficient to separate an object form other structures. If this happens, the user can defined a VOI which prevents segmentation from leaving the area of main interest. To do so, the Restrict to VOIs box has to be enabled and the Define VOI button activated. The VOI tools interface appears and allows drawing a VOI. Outline the VOI. Quit the VOI tools with the OK button to confirm the VOI selection. Make sure the overlay Ovr box is enabled. Finally, activate the Segmentation button to perform the actual segmentation within the VOI. The result is shown in the Mask tab to the right.
To fill in holes present in the mask segment enable the Morphological closing box, select one of the number of pixels available and activate again the Segmentation button.
Saving the Mask for Matching Processing
In order to use a generated segment image as a mask in the matching processing it must be saved as a file. Saving can be performed using the Save Mask pane in any of the supported image formats. Note that automatically the saved file is configured.
Exit the mask interface activating the Close button.
Note that the saved mask is not binary in the case of multiple segments, so that the segments can be recovered. However, during the pixel-wise calculation only the non-zero mask pixels will be processed, while the other pixels are blanked.
2. Load Mask (display definition)
If a mask file already exists, the interactions described above are not necessary and it can be simply configured with the dedicated icon as illustrated below:
It opens a dialog window as shown below:
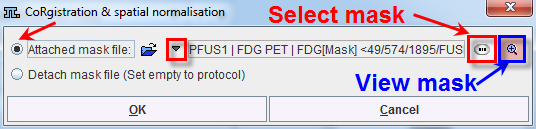
The Attached mask file radio button need to be enabled. The drop down arrow ![]() allows switching between different data formats while the Change file or directory
allows switching between different data formats while the Change file or directory ![]() button can be used to select the mask. The activation of the
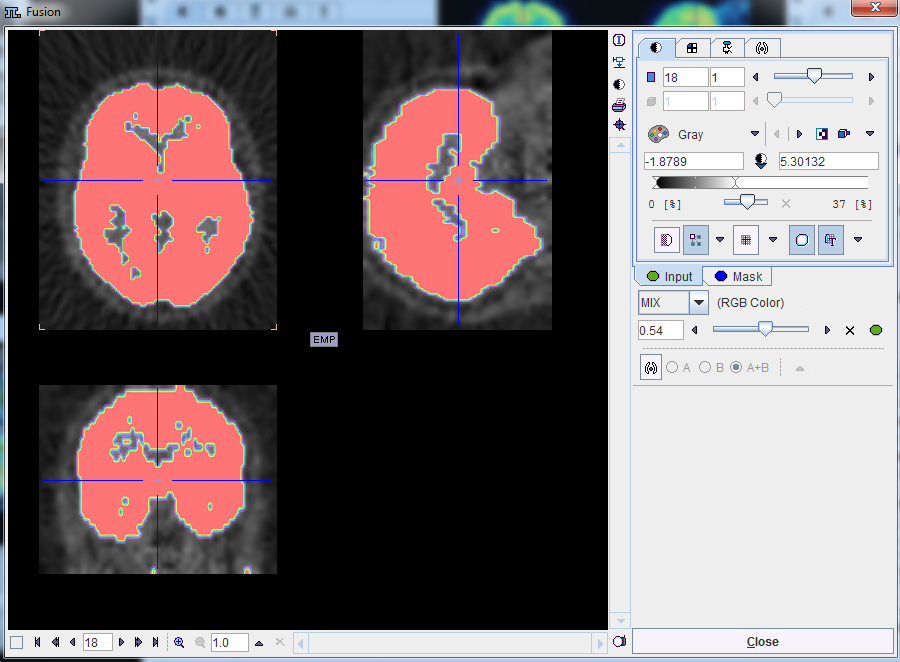
button can be used to select the mask. The activation of the ![]() button next to mask selection allows loading and viewing the mask in a dialog window.
button next to mask selection allows loading and viewing the mask in a dialog window.
Confirm the selected mask to the protocol activating the OK button in the CoRegistration & spatial normalization dialog window. Upon confirmation the window is closed.
Remove the Mask from the Matching Processing
To remove the mask from the matching processing activate the the same dedicated icon from the lateral task bar as illustrated above. A dialog window opens. Make sure the Detach mask file (Set empty to protocol) radio button is enabled and confirm the selection with OK.
3. Mask definition for Automatic Matching
When the masks are defined in the display, they are immediately set as the corresponding Basic parameters to the automatic matching algorithms. Alternatively, the masks can be defined during the setting of the Basic parameters for the automatic procedures. In the example below, the Input mask was defined in the display while no mask was defined for the Reference image. Note the masks section in the Basic parameters settings: a message alerts that for the Input image the Mask defined in image display will be used while for the Reference image the mask can be loaded and set to the protocol. Note that in this case the mask need to be created and saved beforehand. The mask file corresponding to the Input image will allow restricting the matching algorithm to the meaningful area.
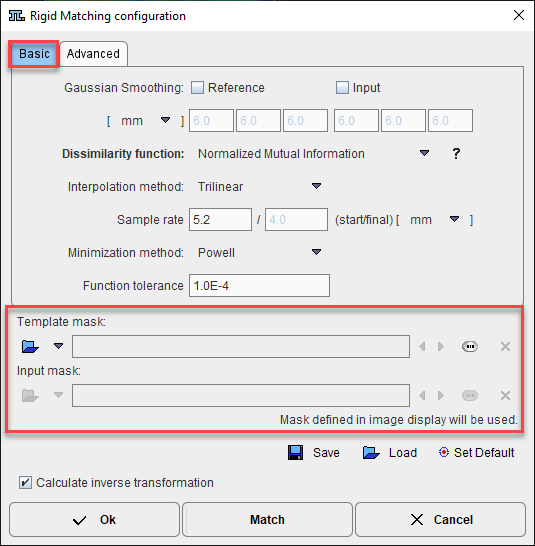
To summarize, there are three potential scenarios:
1.if there is a mask defined in the display it will be set and used during the automatic calculation.
2.if there is no mask defined in the display and there is a mask file defined in the Basic parameters it will be used for the automatic matching.
3.no mask will be used during the automatic matching algorithms if there is no mask defined in the display and no mask defined as a Basic parameter.