Autodetect (Read only)
For users working with different image formats it may be cumbersome to remember the data format of the different files, which may have similar suffixes. The Autodetect format makes it unnecessary to select a specific format before image loading. The user can just select the file(s) he wants to load, and PMOD will try to find out the appropriate format based on the file contents (not on the suffix) when loading.
DICOM (Read/Write)
Image data according to the DICOM standard. As input data, the following Information Objects have been tested: CT, Enhanced CT, MR, Enhanced MR, NM, PET, Enhanced PET, SC, SC multi-frame true color, US.
PMOD's DICOM Conformance Statement describes in detail the DICOM support implemented in PMOD. It can be downloaded from the Support page on the PMOD Website.
Database (Read/Write)
Data loading/saving with a PMOD database. Image data are saved based on the modality: CT as Enhanced CT IOD, MR as Enhanced MR IOD, other images as Enhanced PET IOD.
Query (Read only)
This loading "format" is a shortcut for retrieving images from a remote DICOM server and directly loading them to the image display.
Interfile (Read/Write)
Interfile is a file format developed for data in Nuclear Medicine (Todd-Pokropek A, Cradduck TD, Deconinck F, A file format for the exchange of nuclear medicine image data: a specification of Interfile version 3.3, Nucl Med Commun. 1992 Sep;13(9):673-99).
There are two basic variants of Interfile. One variant has separate header (.hdr) and image (.img) files. This variant has the advantage that the header file can be viewed and edited with a simple text editor. The other variant includes all the information in a single file (.hdr). Both variants are supported in reading and writing, restricted to the subset of data objects which are relevant for PET.
Ecat 6/7 (Read/Write)
Data format of PET data from older CTI/Siemens systems. It is a self-contained format containing all acquisition information.
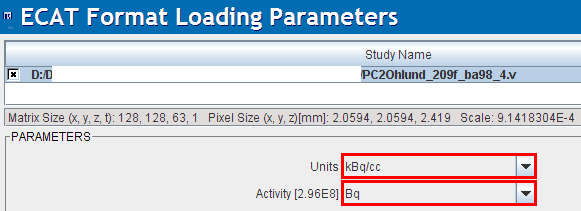
When loading Ecat data an additional choice appears to explicitly define the units of the subject dose values. This facility was introduced because Ecat files without well-defined units were encountered. Experience has shown that the Bq unit should be selected to arrive at the correct doses, but other examples were also encountered. The dose value is important for the SUV calculation and can be checked on the SUV-related panels.
When saving data in the Ecat format, after the specification of the file name, a dialog is presented to the user with a choice of transfer syntaxes. They define the byte ordering used for saving the data. In case you want to read the images using a program other than PMOD, you are recommended to select a big-endian (BE) encoding.
MicroPET (Read only)
Data format of the Siemens Inveon and MicroPET preclinical systems. It is used both for PET and CT data.
NifTI (Read/Write)
Extended variant of the analyze format by the Neuroimaging Informatics Technology Initiative. Both single file (.nii) and double file (.hdr + .img) images are accepted but only in uncompressed form.
PMOD reads the following NIfTI data types:
8-bit unsigned and signed byte (DT_UINT8 and DT_INT8), 16-bit unsigned and signed short (DT_UINT16 and DT_INT16), 32-bit unsigned and signed int (DT_UINT32 and DT_INT32), 32-bit float (DT_FLOAT32), 64-bit double (DT_DOUBLE) and 24-bit RGB (DT_RGB). 64-bit double data are rounded to 32-bit float representation. Up to 4D data is supported, meaning PMOD does not accept files with the 5th dimension greater 1. Only a single value or a single RGB triplet per pixel is supported. If qform_code > 0, PMOD uses qform matrices, otherwise if sform_code > 0 sform matrices will be used. When neither is defined data orientations remain undefined in PMOD. PMOD does not support any extensions nor NIfTI statistical codes. Gzipped images must be uncompressed prior to their usage with PMOD.
NIfTI images produced by SPM require A/P mirroring (TOP to BOTTOM sorting) to get them into HFS position. Other NIfTI images should best be loaded with Reorient to Standard Orientation enabled.
In writing, PMOD supports the variants with two files (header information in *.hdr, image data in *.img) or a composite file (*.nii). The single file variant is required when preparing atlas template images.
Analyze (Read/Write)
Data in the Analyze 7.5 format consists of two files. The header file (*.hdr) contains information about dimensions, pixel representation (transfer syntax) and value scaling. The data file (*.img) contains the pixel data itself without any header offset. Value units and information about acquisition timing must be manually entered, or can be loaded from definition files.
SPM produced Analyze files are usually L/R mirrored. Such files require a 180o rotation about the z-axis s to get them into the radiological head first supine (HFS) position which is the PMOD default.
Note: Analyze has the possibility to include timing information, but only for a single frame. If a timing is specified in the file it will be applied for all frames of a dynamic study, even if a different timing was specified using Edit Times in the user interface. In such cases it is advised to load the image data, change the timing, and save the images again in a format wit better timing support (DICOM, Ecat, Interfile). PMOD writes Analyze files without timing information.
AVW (Read only)
Newer version of the Analyze format.
Raw (Read/Write)
A raw image file which holds only the pixel data. All additional information must be edited manually. Different types of number encoding (byte, signed and unsigned short integer, float) and byte orders (big/little endian) are supported.
Graphic (Read/Write)
Graphic images in jpg, gif and bmp can be read. When writing as a Graphic file, the jpg format is used.
TIFF (Read/Write)
The graphic tiff format is treated separately and can be read and written.
Matlab (Read/Write)
Image data can be read which has been prepared and saved in Matlab using the save -V4 command (Note: only Matlab version 4 format is supported, thus the requirement of the -V4 option). The following Matlab variable names/contents are expected:
▪dim[nx,ny,nz,nTime] (number of columns, rows, slices, acquisitions)
▪vox[xSize,ySize,zSize] (voxel sizes in mm in all 3 dimensions)
▪orig[xOrig,yOrig,zOrig] (position of the origin measured in mm from the top left corner of the first slice)
▪time[1,...nTime] (times when the acquisitions started)
▪timeEnd[1,...nTime] (times when the acquisitions ended)
▪a series of value vectors named v1s1 till v21s35 when nTime=21 and nz=35 (v denotes acquisition number and s slice number)
▪subjectName[] (string with subject name)
▪subjectId[] (string with subject ID)
▪vUnits[] (string with value unit name)
▪tUnits[] (string with time unit name)
Note that only the variables dim and v1s1 etc. are mandatory.
Neurostat (Read only)
Neurostat uses the Interfile format (.hdr, .img) to save the 8 surface projections into an image file. Please note, that this is not a volumetric data set, and therefore should be viewed in a planar layout such as 2x4.
AFNI (Read only)
AFNI (acronym for Analysis of Functional NeuroImages) creates two-file data (.HEAD, .BRIK).
GE Advance (Read only)
Data format of PET data exported from GE PET systems (Advance, Discovery) with the Investigator utility. It is a self-contained format with all acquisition information.
Philips (Read only)
Mosaic data format of PET data from Philips PET (Allegro, Gemini) and older GE Quest systems.
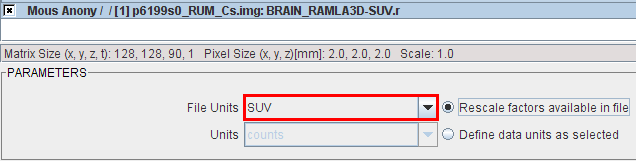
When PMOD recognizes more than one unit defined in the file the loading dialog window shows a choice selection for the user to choose which units will be loaded from a file, providing different range of values.
HIDAC (Read only)
Data format of the (historic) animal PET system HIDAC (Oxford Positron Systems). To be retired.
BrainVISA (Read only)
Format used by the Brainvisa software.
Varian FDF (Read only)
Flexible Image Format of Varian MR systems.