An image series for which a set of contour VOIs has been defined can easily be converted into a VOI atlas as follows:
1.Load the image series in the PVIEW tool.
2.Load the contour VOIs in the VOIs page.
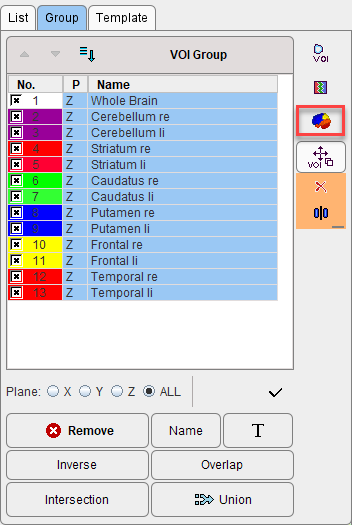
3.On the Group tab, select the VOIs to be included in the atlas.
4.Activate the Create Template Atlas icon on the right side of the VOI Group.
A dialog window appears which requests an Atlas name for template.
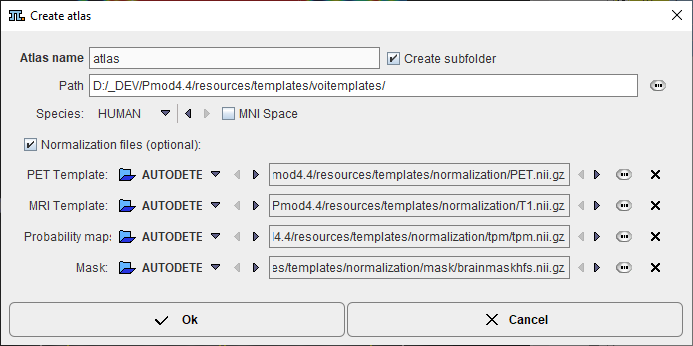
Using this name a subdirectory is created in the Path directory which defaults to resources/templates/voitemplates, but which can be changed. The Species selection allows tagging the VOI atlas so that it will be shown in the corresponding context.
An image series is generated which encodes each VOI pixel as an appropriate numeric label and saved in NifTI format. Additionally, a text file is created for the mapping of the numeric labels to VOI names. The procedure also creates a manifest text file which will contain information about the selected Species: "SPECIES = HUMAN". After activating the refresh button ![]() in the Atlas panel of Template, the new template appears and can be used for statistics.
in the Atlas panel of Template, the new template appears and can be used for statistics.
For human VOI atlases created in standard MNI space it is recommended to enabled the MNI Space box. It creates an entry "SPACE = MNI" in the manifest file which contains information about the species, so that the standard set of normalization templates will be used as normalization files. In such scenario there is no need of definition for the Normalization files (optional).
Important: Because each pixel can only have one numeric label, the VOIs used for the atlas generation may not be overlapping.
Optionally, files for the spatial normalization can be defined for the template which are not in standard MNI space. To this end enable the Normalization files (optional) box and select as many template files as available: The PET Template, MRI Template, and the Mask will be used for the deformable registration. To also use the Probability Maps Normalization a Probability maps file should be selected which encodes the Grey matter, White matter and CSF probability maps as a dynamic data set. In this case, a normalization folder is created with the normalization images in NifTI format. It also contains a mask sub-folder for the mask image (mask.nii), and a tpm folder for the tissue probability map (tpm.nii).