Mouse data
All samples were added to a learning set and the uNET segmentation architecture selected.
Prediction was tested on several additional examples. Preprocessing to rotate new data such that the right ventricle is on left of screen was required for successful prediction. Dark artefacts in the blood can lead to segmentation artefacts that require manual correction before cardiac function can be accurately assessed.
Human data
All samples were added to a learning set and the Multichannel Segmentation architecture selected.
Training was performed as follows:
•notebook Intel i7 8 x 2.3 GHz
•32 GB memory
•no additional cropping
•3225 samples (2580 training, 645 validation)
•106 epochs (aborted)
•batch size 50
•learning rate 0.005
Training took 63 minutes.
Prediction could only be tested on data that was already used in training. Acceptable segmentation results and function analysis was achieved, but the model may be overtrained for this data. The example below shows a result in PCARDM using the MACHINE LEARNING segmentation option that utilises the model trained in this case study.
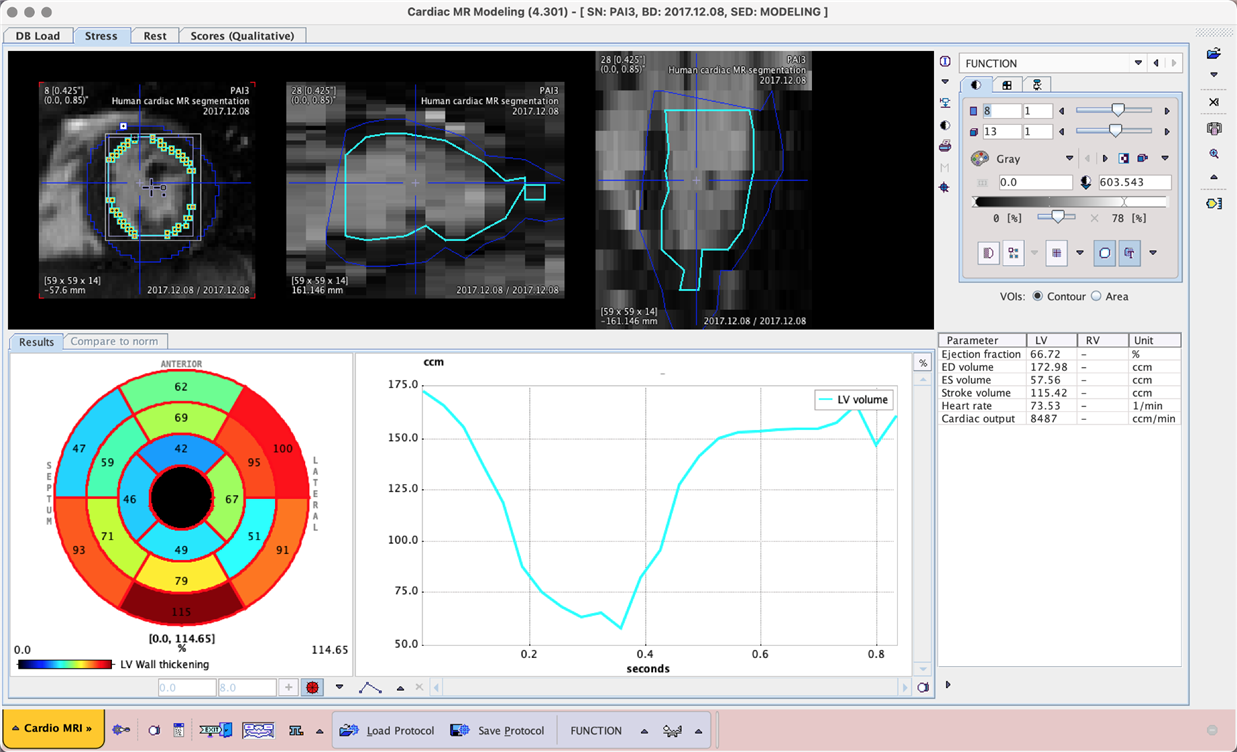
Example data to test the MRI Myocardium 2D and MRI Human Myocardium models is available in our Demo database (Subjects PAI2 and PAI3). The data used for the case study was provided by Bruker colleagues and in a private collaboration.
To try the models for yourself we recommend use of the Cardiac MR Modeling tool (PCARDM) FUNCTION workflow. See the specific documentation for PCARDM. The model selection is determined by the species selection in PCARDM. The models were trained with 2D data so Split Slices and Split Frames are required. The recommended Heart Box for example PAI2 is 10 x 10 mm, and for PAI3 100 x 100 mm. The data used in these cases was for a limited range of MR sequences - the performance of the model may vary for data from different hardware and/or with different contrast/pre-processing.