Many studies have been performed using amyloid PET in the search for a biomarker predicting progression to ALzheimer’s/dementia, and several databases of imaging data for amyloid PET tracers are available online (often along with anatomical MR and FDG PET). Visually, there are pronounced differences between the distribution of the amyloid PET tracer 11C-PiB in subjects without significant amyloid accumulation in cortical regions and in subjects with large accumulation. In subjects without significant accumulation, tracer uptake is clear in white matter with very low signal in grey matter/cortical regions. In subjects with significant amyloid accumulation the tracer is more uniformly distributed throughout the brain with grey matter uptake approaching/equalling white matter. Note that healthy subjects may still have amyloid accumulation and amyloid accumulation is not clear in all cases of Alzheimer’s that undergo imaging.
We used an online database of 11C-PiB PET to illustrate the functionality of our AI classification using the SVM architecture:
http://www.gaain.org/centiloid-project (Standard PiB data)
This database contains amyloid PET data for subjects with Alzheimer’s disease (AD) and amyloid accumulation, and comparative data from young controls (YC) with low/negligible amyloid accumulation.
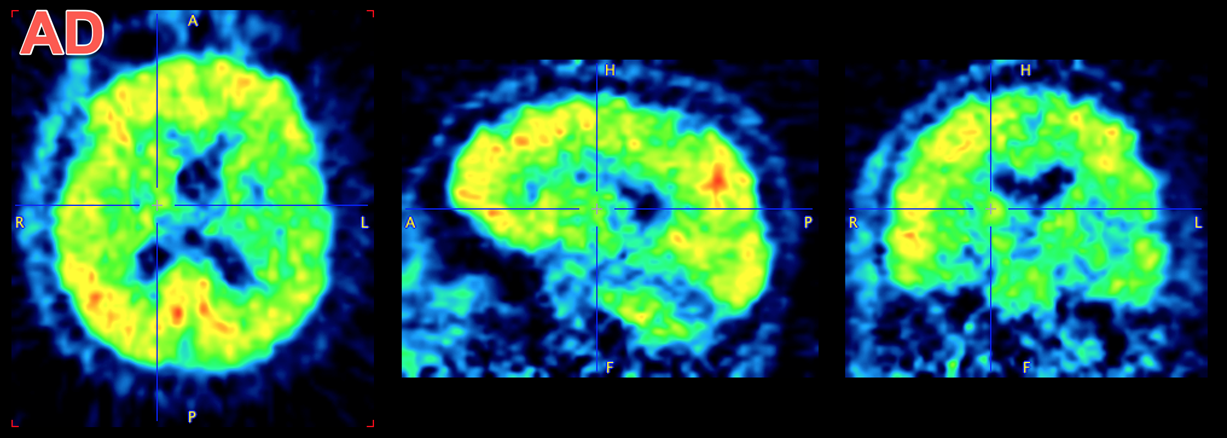
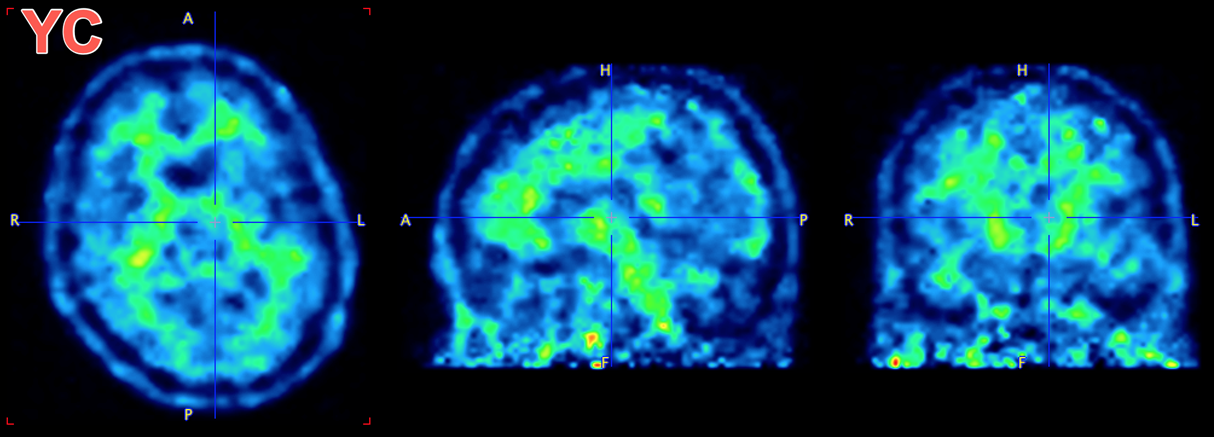
Example data to test the Amyloid PET SVM model is available in our Demo database (Subject PAI7). The data used for the case study was downloaded from the GAAIN project website: http://www.gaain.org/centiloid-project
To try the model for yourself the workflow in the View tool is required. If new data is tested, cropping should be applied to reduce the field-of-view to the brain. The model selected should be AmyloidPET-SVM (Classification SVM). The data used in these cases was all cropped 3D PET for late average 11C-PiB from unknown PET hardware - the performance of the model may vary for data from different hardware and/or with different tracers/reconstruction/pre-processing.