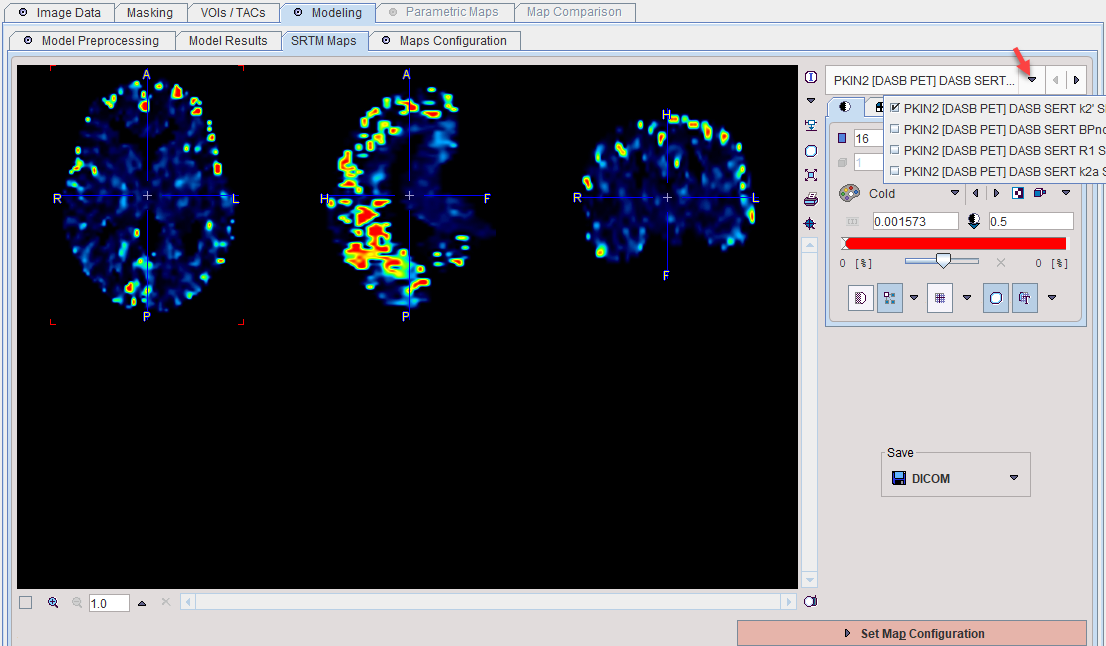
Commonly, reference tissue models are based on the following structure, as described in more detail here.
The k2' parameter describes tracer clearance from the reference tissue. Some of the reference tissue models estimate k2' in each individual pixel, whereas it actually should be a common constant. There are several reference tissue models which leverage this physiologic constraint and require a fixed k2' as input. There are different approaches to obtain an adequate k2' value as described below.
k2' Specification in Model Preprocessing Panel
Models which require a k2' input support four ways of specifying k2' reflected in Model Preprocessing panel by a radio button selection.
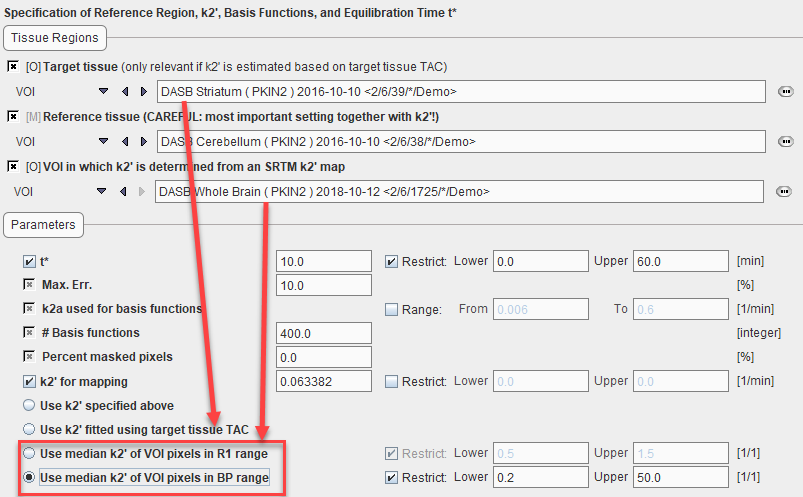
1.Use k2' specified above: Use of an externally determined value which is entered manually in the k2' for mapping field. In PKIN, two approaches for getting such a k2' can be implemented. First, regional TACs are calculated from the dynamic PET and loaded into PKIN. One of the TACs should represent appropriate reference tissue (without receptors), and the others tissue with high specific binding. The latter TACs are then fitted with one of the reference models yielding k2' as an output (MRTM, SRTM), and the resulting k2' values averaged. Alternatively, the SRTM2 model can be used and coupled fitting performed, with k2' being estimated as a common parameter. More detail on these approaches is available in the PKIN Users Guide.
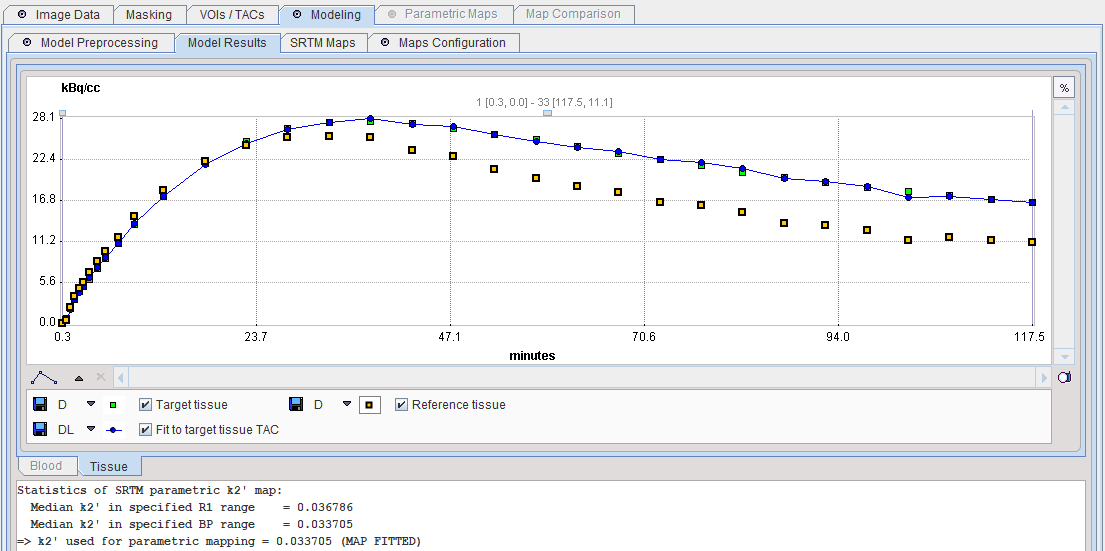
2.Use k2' fitted using target tissue TAC: During preprocessing k2' is fitted by applying the SRTM model to the specified Target tissue and Reference tissue curves. The estimated value is reflected in the k2' for mapping field.
3.Use k2' of VOI pixels in R1 range: During preprocessing the SRTM model is applied to calculate k2' and R1 maps in a specified sub-volume avoiding the reference tissue. The median value of all k2' values within a physiologic R1 range represents the estimated k2' value, which is reflected in the k2' for mapping field.
4.Use k2' of VOI pixels in BP range: During preprocessing the SRTM model is applied to calculate k2' and BPnd maps in a specified sub-volume avoiding the reference tissue. The median value of all k2' values within a physiologic R1 range represents the estimated k2' value, which is reflected in the k2' for mapping field.
k2' Using the two Parametric Mapping Approaches
The two methods described above using SRTM for parametric mapping of k2' are illustrated using the SRTM2 model with the preprocessing parameters
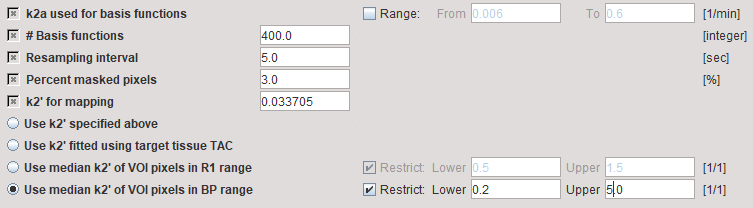
The Model Results panel shows the SRTM fit to the Target tissue and the results of k2' parametric mapping. Both median variants are calculated, and the BP-restricted value will be used for parametric mapping.
An additional SRTM Maps panel is available to actually inspect and save the maps.