The MRGlu (FDG Patlak) model is intended for the quantitative assessment of the regional metabolic rate of glucose (MRGlu) with FDG. The required measurements are a dynamic PET scan after the injection of a FDG bolus and external blood sampling. The analysis is done using the Patlak graphical plot method [1] which has been developed for systems with irreversible trapping, ie. k4=0 in a 2-tissue compartment model.
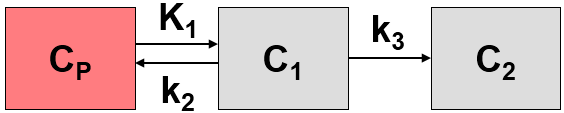
However, this model structure is not necessary for the application of the method. It is sufficient to have any compartment in the system which binds irreversibly.
The Patlak plot belongs to a group of Graphical Analysis techniques, whereby the measured tissue TAC CT(T) undergoes a mathematical transformation and is plotted against some sort of "normalized time". The Patlak plot is given by the expression
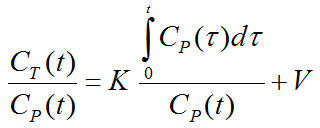
with the input curve Cp(t). This means that the measured PET activity is divided by plasma activity, and plotted at a "normalized time" (integral of the input curve from the injection time divided by the instantaneous plasma activity). For systems with irreversible compartments this plot will result in a straight line after an equilibration time t*.
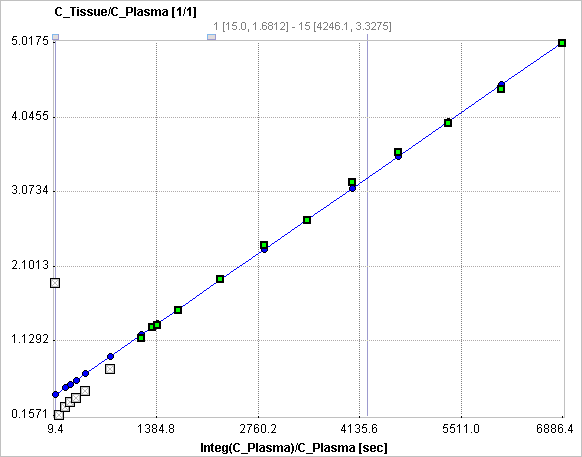
The slope K and the intercept V must be interpreted according to the underlying compartment model. For the FDG tracer, the slope K equals K1k3/(k2+k3) and represents the metabolic flux, while the intercept V equals V0+vB with the distribution volume V0 of the reversible compartment C1 and the fractional blood volume vB.
For the analysis of FDG data, the Lumped Constant (LC) and the Plasma glucose level (PG) of the patient need to be entered. The metabolic rate of glucose MRGlu is then obtained from the regression slope K by
![]()
Acquisition and Data Requirements
Image Data |
A dynamic PET data set representing the measurements of brain activity from the time of injecting of a 18F-Deoxy-Glucose (FDG) bolus. |
Input curve |
Plasma activity of blood sampled at a peripheral artery from the time of injection until the end of the acquisition. |
Target tissue |
A regional time-activity curve from a brain region. It is presented as a Patlak plot and can be used to define the linear segment where the regression analysis should be done. |
Blood Preprocessing
Decay correction is the only blood correction option. Note that an uncorrected relative time shift of blood data by 30 sec does not markedly change the calculated glucose consumption.
Model Preprocessing
The Patlak graphical plot is performed with the TAC from the specified tissue VOI and presented to the user. Essentially, it is a plot of the TAC with "normalized time" along the x-axis and "normalized" tissue activity on the y-axis. In this plot, the TAC becomes linear after an equilibration time due to irreversible trapping (k4=0). The slope of the linear segment equals the influx constant Ki and can be used to calculate metabolic rate of glucose. The user must decide on the begin of the linear segment in the "normalized" time units and specify the corresponding acquisition start time t* in the model configuration. An alternative is to apply the automatic criterion Max. Err. for fitting t*.
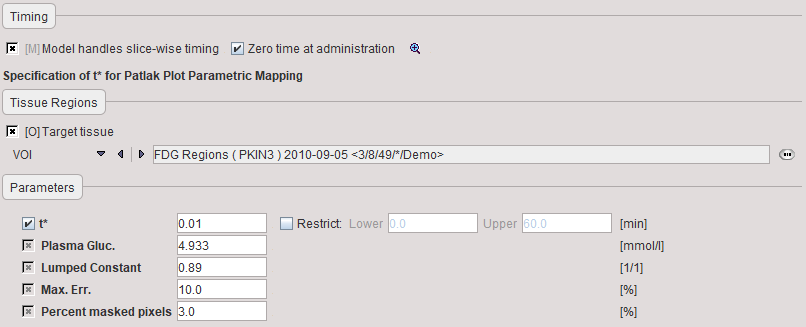
t* |
The linear regression estimation should be restricted to a range after an equilibration time. t* marks the beginning of the range used in the multi-linear regression analysis. It can be fitted based on the Max. Err. criterion. Note that the t* is in acquisition time. |
Plasma glucose |
Plasma glucose in [mmol/l] measured with a blood sample of the patient. |
Lumped constant |
The Lumped constant is used to compensate for the difference in uptake between normal glucose and Fluoro-Deoxyglucose (FDG). Proposed as the default is the value determined by Graham et al (J Nucl Med 2002; 43:1157–1166) using 11C Glucose and FDG in 2002. Their results: Normal brain: 0.89+/-0.08; cerebellum: 0.78+/-0.11. |
Max. Err. |
Maximum relative error ( (measured-predicted)/predicted ) allowed between the linear regression and the Patlak-transformed measurements in the segment starting from t*. |
Percent masked pixels |
Exclude the specified percentage of pixels based on histogram analysis of integrated signal energy. Not applied in the presence of a defined mask. |
If a target tissue was specified, the Patlak plot is shown in the preprocessing Result. The user should consult this plot in order to check whether the t* time is adequate. Otherwise the panel remains empty.
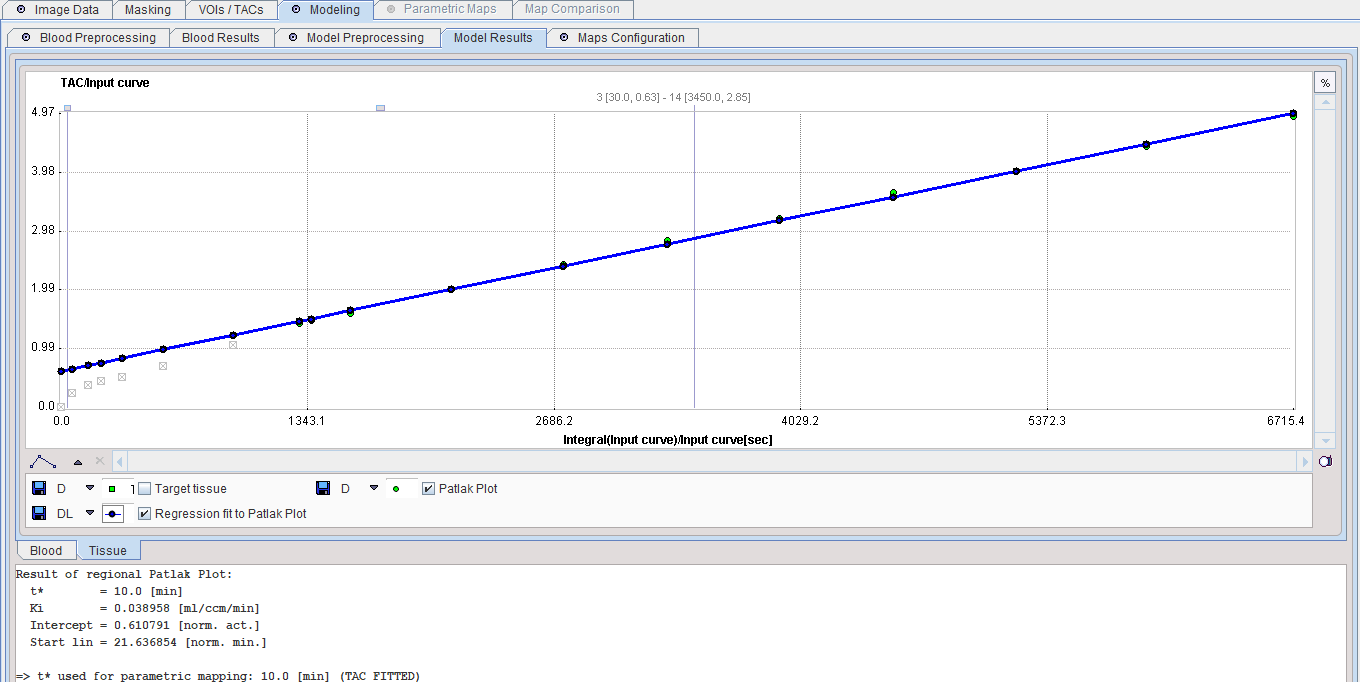
Model Configuration
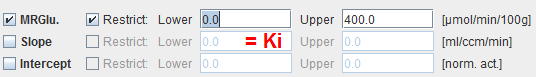
MRGlu |
Metabolic Rate of Glucose in [μmol/min/100g], the actual result of the model. It is calculated as: |
Slope |
Slope of the linear regression. It equals the influx Ki=(K1*k3)/(k2+k3) of the 2-tissue compartment model and is directly proportional to MRGlu. |
Intercept |
Intercept of the linear regression. |
Reference:
1.Patlak CS, Blasberg RG, Fenstermacher JD: Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J Cereb Blood Flow Metab 1983, 3(1):1-7. DOI