Overview
PMOD provides templates of human brain VOIs in the standard MNI space. The PVC method described below combines this information with the grey and white matter masks derived from an individual subject MR image as follows:
1.The anatomical MRI of the subject is segmented and the GM and WM masks calculated.
2.The MR image is normalized to the MNI MR template using the Brain Normalization method.
3.The VOI template is transformed to the MR space using the inverse of the normalization transform.
4.The transformed VOI template is intersected with the GM mask to obtain the GM VOIs in the MR space.
5.The WM mask is converted into a WM VOI.
6.The PET images are rigidly matched to the MR images.
7.The GM and WM VOIs are transformed to the PET space using the inverse rigid transform.
8.A contouring procedure is applied to get contour definitions of all VOIs, which the user can view and edit.
9.The GTM PVC method is applied to the original PET series using this set of template-based VOIs.
Starting the PVC
As a first step, load the PET images into the PVIEW tool. Then activate the ![]() button to the right of the image and select the PVC (VOI based) method from the list of external tools. A dialog window is shown which allows performing partial-volume correction in a step-by-step mode or as a background process.
button to the right of the image and select the PVC (VOI based) method from the list of external tools. A dialog window is shown which allows performing partial-volume correction in a step-by-step mode or as a background process.
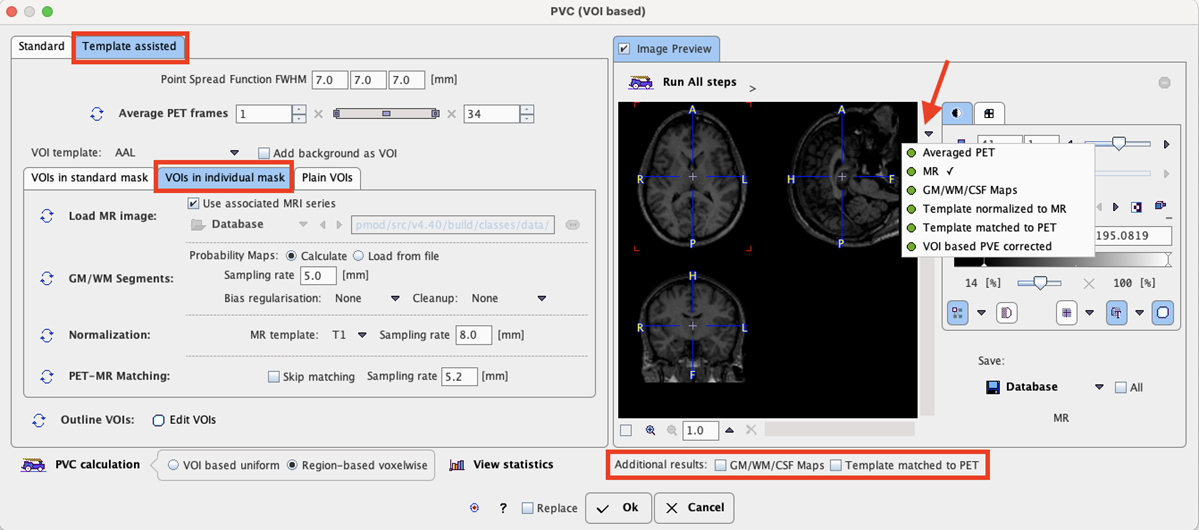
Step-by-Step PVC Processing Mode
The step-wise mode is activated by checking the box next to the Image Preview label. Note the ![]() buttons which are used to start the individual processing steps. Initially, several of them are inactive because the prior steps are missing.
buttons which are used to start the individual processing steps. Initially, several of them are inactive because the prior steps are missing.
The result images of the different steps are collected in the Image Preview area. In the image selection list, available results appear with a green mark, while yet unavailable ones are marked in red. The example above shows the MR loaded by step Subject's MR image. The images selected in the Image Preview can be exported for later use by the Save button.
The advantage of step-wise processing is that steps can be repeated with different parameters until the outcome is satisfactory.
Point Spread Function FWHM:
The PSF is assumed to be a three-dimensional Gaussian function. The FWHM values in the three directions have to be specified according to the resolution of the reconstructed image. Default is 7 mm isotropic FWHM.
Average PET frames:
Dynamic images can be processed and will result in a corrected dynamic series. However, for the PET-MR Matching step, a static PET image showing anatomical information is required. Therefore, the user should define a suitable range for averaging time frames and then activate ![]() . In the case of a static scan this step is not required and the button therefore not active. The Averaged PET is shown in the Image Preview window.
. In the case of a static scan this step is not required and the button therefore not active. The Averaged PET is shown in the Image Preview window.
Subject's MR image:
The user must specify an anatomical MR image of the same subject which will be segmented. He can choose the format, select the image and then activate ![]() for loading the MRI.
for loading the MRI.
![]()
The Use associated MRI series box allows taking advantage of the feature that two series can be associated as a pair in PMOD databases.
GM/WM Segmentation:
This step needs to provide the different tissue segments. If Probability Maps is configured as Calculate, the ![]() button calls a segmentation procedure which may take several minutes to complete. The procedure has three parameters: Sampling rate determines the density of pixels considered in the calculation. Bias Regularisation serves for compensating modulations of the image intensity across the field-of-view. Depending on the degree of the artifact, a corresponding setting can be selected from the list. Cleanup is a procedure for rectifying the segmentation along the boundaries. It is recommended to use the default settings and only experiment with other parameter values if the segmentation fails.
button calls a segmentation procedure which may take several minutes to complete. The procedure has three parameters: Sampling rate determines the density of pixels considered in the calculation. Bias Regularisation serves for compensating modulations of the image intensity across the field-of-view. Depending on the degree of the artifact, a corresponding setting can be selected from the list. Cleanup is a procedure for rectifying the segmentation along the boundaries. It is recommended to use the default settings and only experiment with other parameter values if the segmentation fails.
The results are three segments, WM, GM and CSF. They are arranged as frames in a "dynamic" series. In the illustration below, the three frames are arranged in three columns. Because the calculation takes long, it may be helpful to save the segment images for later use by selecting them in Image Preview and then using the Save button.
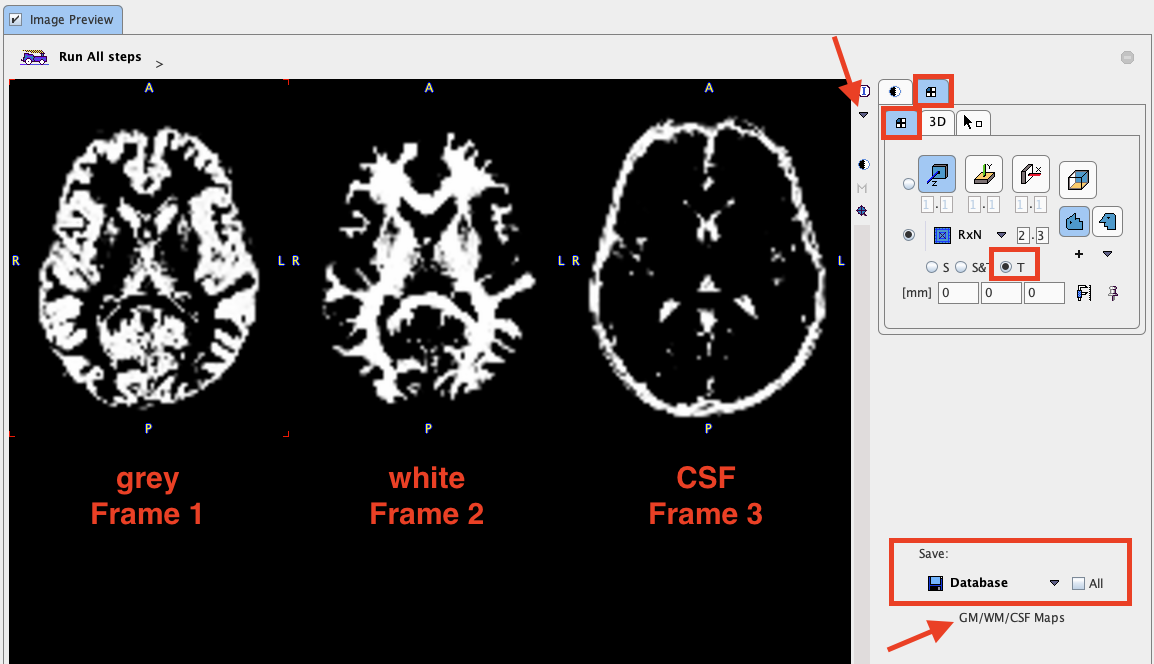
If segment images are already available, Probability Maps can be configured as Load from file, and the corresponding segment file selected. Note that the segment images must be matched to the MR image and different segments should appear as dynamic frames in the indicated order. In this case the ![]() button just loads the segments.
button just loads the segments.
Normalization:
The normalization of the MR image to the MNI template has two parameters, the MR template which should be set to T1 or T2 as appropriate, and the Sampling rate. After calculating the normalization its inverse is applied for transforming the VOI template to the MR space. It is then intersected with the individual GM mask, and the WM mask is used to create the WM VOI. The resulting VOI template is shown in the Image Preview window as Template normalized to MR .
PET-MR Matching:
Upon activating the ![]() button, the PET image is rigidly matched to the selected MR image. The inverse transformation is applied for transforming the VOI template from the MR to the PET space. The result Template matched to PET is shown in the Image Preview window.
button, the PET image is rigidly matched to the selected MR image. The inverse transformation is applied for transforming the VOI template from the MR to the PET space. The result Template matched to PET is shown in the Image Preview window.
If the PET and MR images are already matched, the Skip Matching box can be checked to skip this processing step. However, please note that in this case the MRI and the PET images must have identical pixel size and image matrix. If the automatic matching in the PVC tool is not feasible, matching can be performed manually in the fusion tool and the results saved for use in the PVC tool.
Outline VOIs:
This step calculates contour VOIs from Template matched to PET and shows them together with the Averaged PET in the Image Preview window. This is the VOI set which will be used for PVC.
Edit VOIs:
This button opens a VOI dialog window showing the VOIs on top of the Averaged PET. Note the extended White Matter VOI which has been derived from the WM mask. The VOIs can be inspected and edited, or saved for later use. To return modified VOIs, close the window with the Ok button, otherwise Cancel.
PVC Calculation:
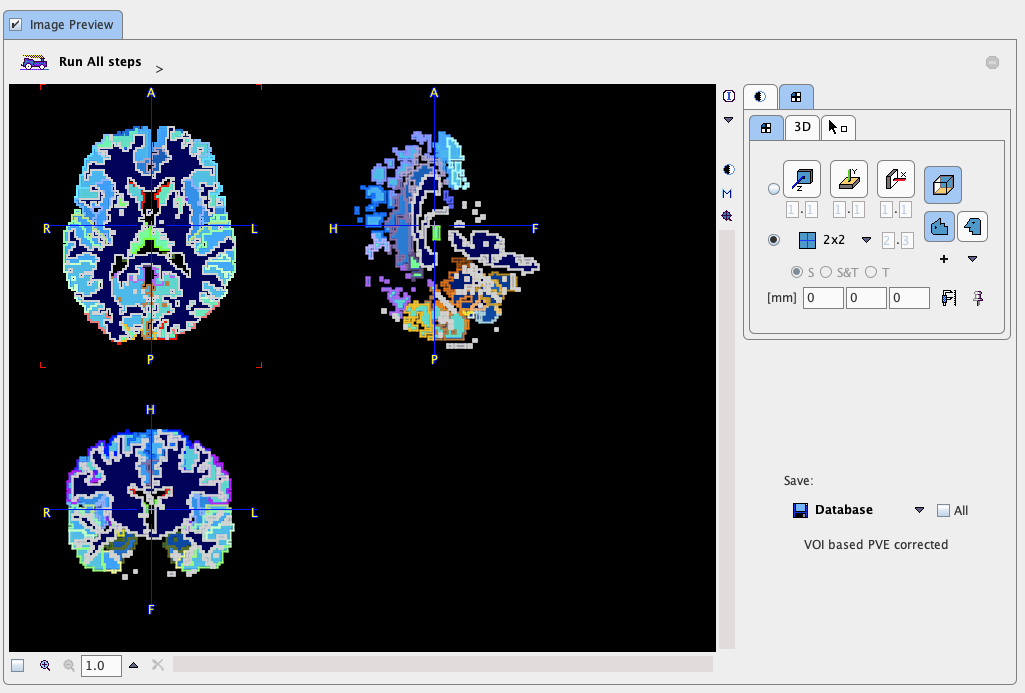
This step performs the actual PVC calculation. Because of the large number of VOIs in the standard templates the processing takes a while. The result is returned in the form of an image series and shown in the Image Preview window. If the input series was dynamic, the result is also dynamic as in the example shown below.
View Statistics:
This button calculates the VOI statistics in the original and the PVC corrected images. Depending on the input images the results are simple statistics, or tissue time-activity curves.
Returning the Results:
Make sure that the Additional results of interest are checked. Then close the window with the Ok button to return the results.
Background PVC Processing Mode
If no interactive processing is desired, the user interaction is minimal.
1.If desired, edit any of the parameters, or reset them by the ![]() button.
button.
2.If the PET series is dynamic, define an appropriate frame range for averaging.
3.Select the anatomical MRI of the same subject.
4.Make sure that the Additional results of interest are checked.
5.Start the PVC calculation with the Ok button.
The dialog window will be closed and processing will run in the background. Once the result were calculated and returned to PVIEW, a confirmation message will be shown.