A Learning Set was created and the Classification SVM architecture selected. The linear kernel was used with a maximum 32 iterations and Cost factor = 1.0.
Training was performed as follows:
•Windows workstation Intel i7-2600K 3.4GHz (no GPU use possible for SVM)
•24 GB RAM
•79 samples (45 class = AD, 34 class = YC)
•The samples were Resized to Box size 160 x 160 x 160 mm and Voxel size 2.0 x 2.0 x 2.0 mm, resulting in a new standardized image matrix of 80 x 80 x 80 for all samples.
•The image values were normalized using the z-score method
•Image reduction was applied using MIP (maximum intensity projection)
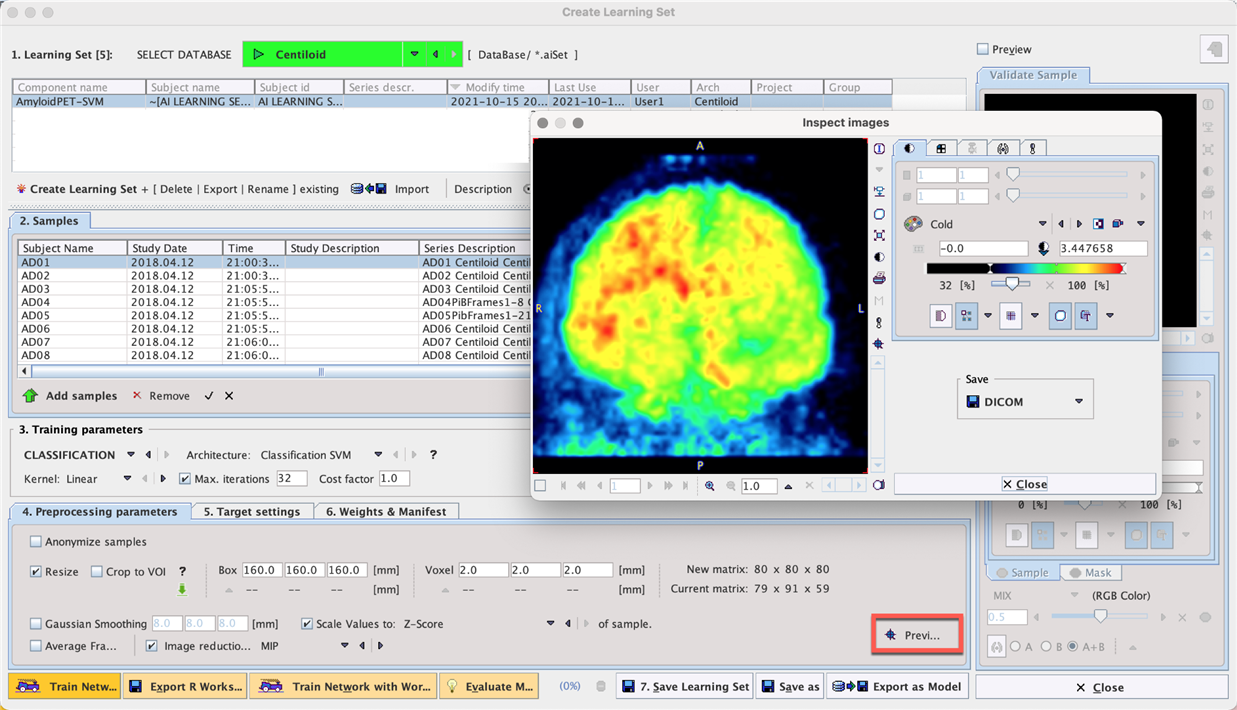
Training took less than 5 minutes.
Randomly selected AD and YC samples were selected from the same dataset to test Prediction. The 3D input data is loaded and the MIP is calculated as part of classification. High probabilities for the expected class were returned.
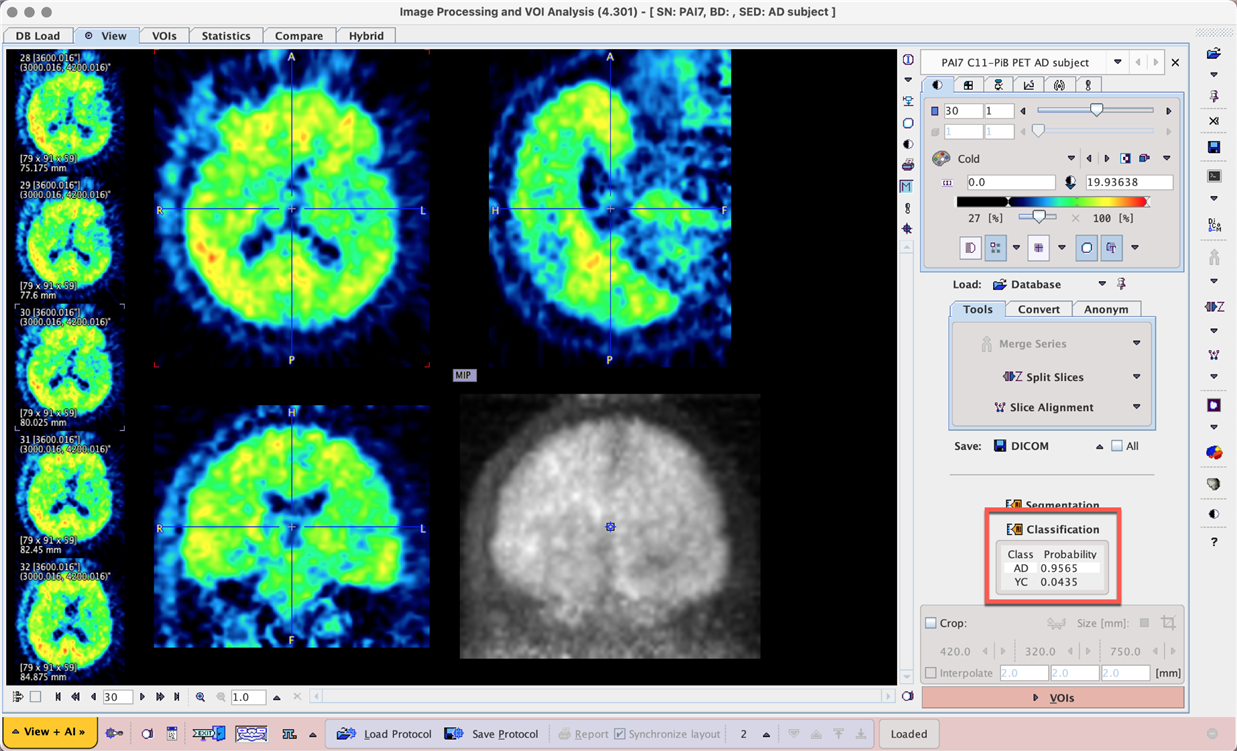
Example data to test the Amyloid PET SVM model is available in our Demo database (Subject PAI7). The data used for the case study was downloaded from the GAAIN project website: http://www.gaain.org/centiloid-project
To try the model for yourself the workflow in the View tool is required. If new data is tested, cropping should be applied to reduce the field-of-view to the brain. The model selected should be AmyloidPET-SVM (Classification SVM). The data used in these cases was all cropped 3D PET for late average 11C-PiB from unknown PET hardware - the performance of the model may vary for data from different hardware and/or with different tracers/reconstruction/pre-processing.