The MRGlu (FDG Autorad) model is intended for the quantitative assessment of the regional metabolic rate of glucose (MRGlu). The required measurements are a static PET scan between 40 and 55 min after the injection of a FDG bolus, and external blood sampling from the time of injection until the end of the PET acquisition, as well as the analysis of one blood sample for the plasma glucose concentration. The analysis is based on an autoradiographic solution of the 2-tissue compartment model. It provides an operational equation (eq. (8) in [1]) as to how calculate the metabolic rate of glucose from the blood curve, the PET value, the plasma glucose, and 5 fixed (but modifiable) parameters: the lumped constant, K1, k2, k3, and k4.
Acquisition and Data Requirements
Image Data |
A static PET data set representing the measurement of the average brain activity between 40 to 55 min after injection of a FDG bolus. If a dynamic study is loaded, the activity of all loaded frames is averaged at the time of pixelwise model calculation. |
Input curve |
Plasma activity of blood sampled at a peripheral artery from the time of injection until the end of the acquisition. |
CAUTION:
The timing and decay correction of the blood and image data has to be synchronized. It is recommended to use times relative to the injection of the tracer. So the blood sampling times and the decay correction should be relative to injection time. For a static PET scan decay correction is usually performed to the acquisition start. Therefore, in order to correct to the injection time, the image has to be scaled accordingly. Furthermore, the image acquisition start and end times have to be set relative to the injection time.
In practice it is recommended loading the PET image in the viewing tool, scaling it, editing the acquisition timing, saving it in a format supporting proper timing (DICOM, Ecat, Interfile), and using that file for the PXMOD analysis.
Blood Preprocessing
Decay correction is the only blood correction option. Note that an uncorrected relative time shift of blood data by 30 sec does not markedly change the calculated glucose consumption.
Model Preprocessing
The rate constants applied in the autoradiographic calculation of MRGlu must be entered in the Model Preprocessing dialog.
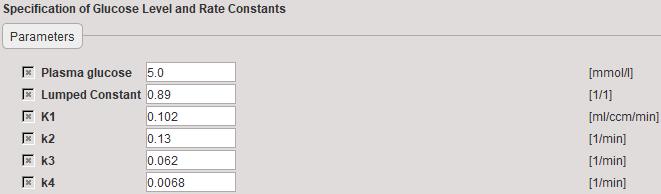
Plasma glucose |
Plasma glucose in [mmol/l] measured with a blood sample of the patient. |
Lumped constant |
The Lumped constant is used to compensate for the difference in uptake between normal glucose and Fluoro-Deoxyglucose (FDG). Proposed as the default is the value determined by Graham et al (J Nucl Med 2002; 43:1157–1166) using 11C Glucose and FDG in 2002. Their results: Normal brain: 0.89+/-0.08; cerebellum: 0.78+/-0.11. |
K1 |
Unidirectional transfer of FDG into tissue. Grey matter: 0.102. White Matter: 0.054. |
k2 |
Clearance of FDG from the tissue. Grey matter: 0.130. White Matter: 0.109. |
k3 |
Phosphorylation rate in tissue. Grey matter: 0.062. White Matter: 0.045. |
k4 |
Dephosphorylation of glucose-phosphate in tissue. Grey matter: 0.0068. White Matter: 0.0058. |
Model Configuration
![]()
MRGlu |
Metabolic Rate of Glucose in [μmol/min/100g] calculated according to eq. (8) in [13]. |
Reference
1.Huang SC, Phelps ME, Hoffman EJ, Sideris K, Selin CJ, Kuhl DE: Noninvasive determination of local cerebral metabolic rate of glucose in man. The American journal of physiology 1980, 238(1):E69-82.