A VOI atlas in PMOD consists of the following components:
1.Atlas image: Image which encodes the atlas VOIs in a stereotactic space as numeric labels.
2.Label list: Text file mapping the label values to the VOI names shown in the user interface.
3.Manifest: Text file that defines the properties of the atlas.
4.Normalization files for calculation of the transformation between the subject anatomy and atlas anatomy (not needed for MNI atlases).
The atlas information has to be organized in a sub-directory of resources/templates/voitemplates exactly as illustrated below for the AAL-Merged atlas.
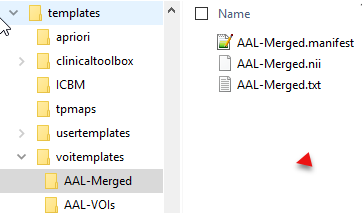
The atlas name (e.g. AAL-Merged) has to be used as the sub-directory name, the atlas image name (AAL-Merged.nii), the label list name (AAL-Merged.txt), and the manifest name (AAL-Merged.manifest). If the atlas is not a human atlas in the MNI space, it needs to include an additional normalization folder for the templates as described below.
By conforming to this structure it is possible for users to prepare their own VOI atlases. In PNEURO, all atlases can be used for the Maximum Probability Atlas approach, whereas the Brain Parcellation approach only supports human atlases in the MNI space.
Atlas Image
The atlas image must be prepared as a NifTI file and encode the atlas VOIs as numeric labels. Each pixel has a value of 0 if it is a background pixel, or otherwise an integer number. We recommend using the HFS anatomical orientation (head first, supine = radiological convention) for human data.
Label List File
The label list text file has the minimal form:
name1 outlined_name1 label_value1
name2 outlined_name2 label_value2
where each VOI is represented by a line.
The list can be extended with additional information for the VOI presentation as illustrated below for the AAL-Merged.txt. The first column starts with the name followed by a bracket construction which encodes a tree structure. For instance, Precentral belongs to the Frontal_lobe which is located in the left Left or right Right_hemisphere of the Cerebral_cortex. The second column indicates the name of a generated contour VOI. The third column contains the label value in the atlas file. Each pixel in AAL-Merged.nii with value 1 will belongs to the Precentral_l VOI, pixels with value 2 to Precentral_r, etc. The third column specifies the RGB color values for showing the VOI, and the last column the text to be shown as a tooltip.
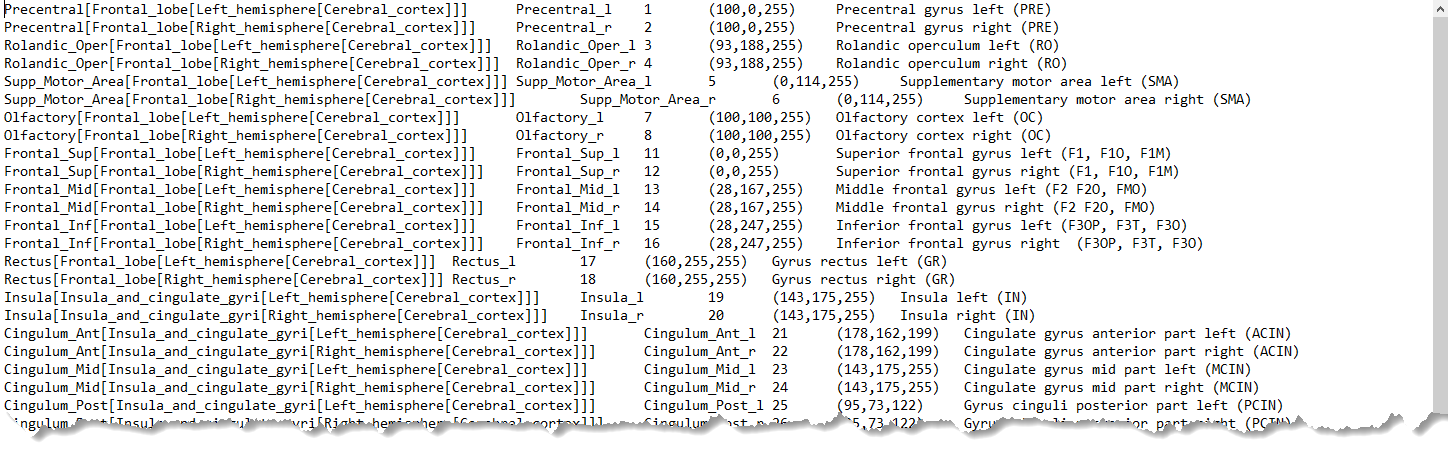
Note: For all rows, all columns need to be filled. Spaces in the Name field are not allowed.
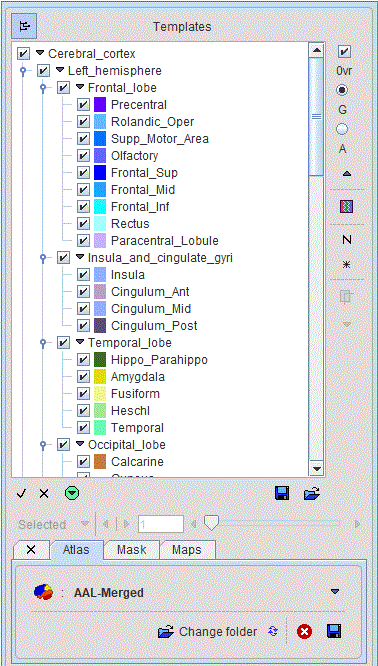
The corresponding atlas VOI structure is illustrated below.
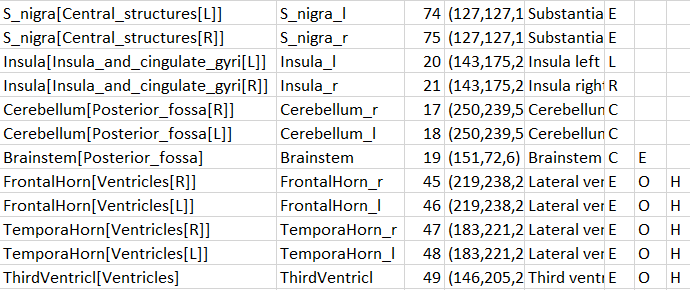
There are additional options to be added to the columns for use in PNEURO:
•E: Excluded from masking by the grey matter threshold.
•O: Indicates that the VOI is not brain matter.
•H: Indicates that the VOI should initially be hidden, i.e. not selected on the Group panel.
•L, R: Indicates that the VOI belongs to the left (L) or right (R) hemisphere. This information is used in sulci deformation.
•C: Indicates that the VOI belongs to the cerebellum. This information is also used in sulci deformation.
Example as shown in Excel:
Manifest File
The following entries are supported in the manifest text file describing the atlas:
SPECIES = HUMAN |
The supported species include HUMAN, PRIMATE, PIG, RAT, MOUSE |
SPACE = MNI |
Only the MNI (Montreal Neurological Institute) space for humans is supported. If this line is present, a common set of template files with 2x2x2mm resolution is used. |
APPLICATION = NO_PNEURO |
If this line is present, the atlas is not listed in PNEURO. |
TYPE = PROBABILISTIC, RANGE 0 / 100 |
The range specified for the data will be scaled to a probability value in the range [0,1] |
Spatial Normalization Methods
Atlases can only be applied to images if they have the same resolution and show the anatomy with the same geometry. Therefore, images originating from real experiments first need a normalization step for the atlas to be applied. This is done by calculating a normalization transform between the subject image and a "template" image representing the standard anatomy imaged with a certain modality, and using it for warping the VOIs to the subject anatomy.
Name of method in PMOD |
Methodology and template |
Template-based normalization |
Implementation of the spatial normalization in SPM5. It requires a template image in the atlas space which has a similar pattern as the image to normalize. The usual reference images are the PET, T1 and T2 SPM5 templates. The appropriate template has to be selected by the user. |
3 Probability maps normalization |
Implementation of the unified segmentation normalization in SPM8. It requires tissue probability maps of grey matter, white matter and CSF in the atlas space, which is organized in a dynamic file called tpm.nii. This normalization is mostly applicable to T1-weighted MR images. |
6 Probability maps normalization |
Implementation of the unified segmentation normalization in SPM12. It uses three additional probability maps (bone, tissue and air), which are organized in a dynamic file tpm_6.nii. This normalization is mostly applicable to T1-weighted MR images. |
Template Files for Template-based Normalization
For atlases of the human brain anatomy which are defined in the 2x2x2mm MNI space and which have the entry SPACE = MNI defined in the manifest file, a common set of template files for the Template-based normalization is available in the resources/templates/normalization folder.
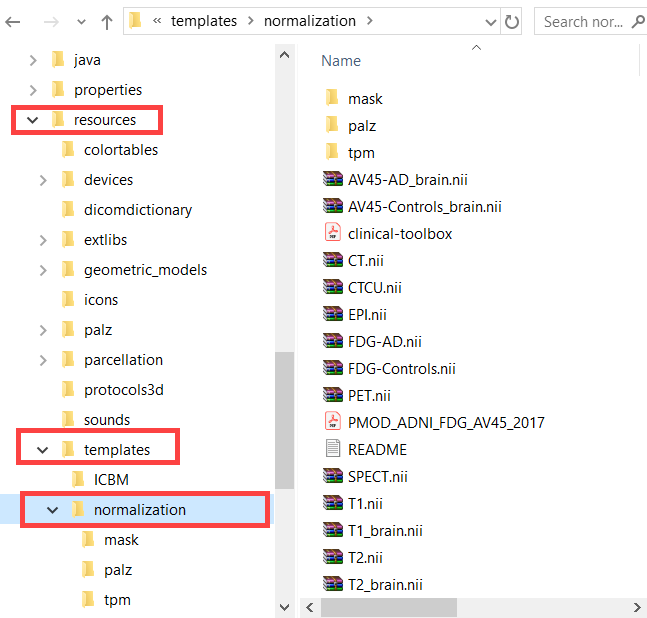
For atlases in other spaces than MNI corresponding normalization templates have to be included in a normalization sub-folder. As an example, for the Cynomolgus (CIMA_UN) atlas illustrated below FDOPA, DTBZ and an MRI template are available.
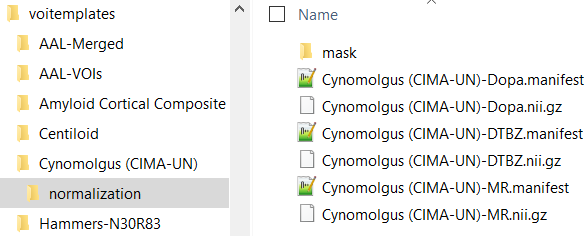
In the example above a manifest file with species definition is available for each normalization template.
Normalization works best if the information is restricted to the relevant image part. Therefore, normalization should contain a mask sub-folder with a mask file mask.nii containing 1 for all relevant pixels and 0 for all others.
Resource Files for Probability Normalization Methods
Again, for a human atlas defined in the MNI space the tissue probability maps are already available in the templates/normalization/tpm directory.
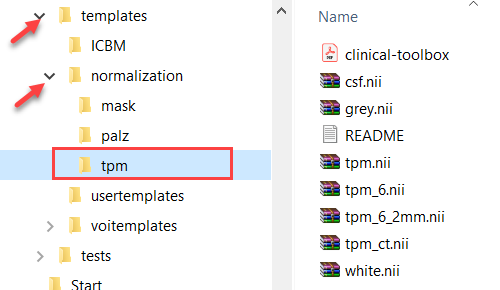
To use the methodology for other than MNI atlases, a normalization/tpm sub-directory has to be added to the atlas directory with the corresponding tissue probability maps.