If a labeled metabolite of the tracer enters tissue, the additional signal has to be accounted for in the model. The current model introduced by Fujita et al. [1] includes a second input curve CM(t) of a metabolite entering tissue and undergoing non-specific binding. C1 represents the non-displaceable compartment of the authentic ligand, C2 the specific binding of interest, and C3 metabolized ligand in tissue.
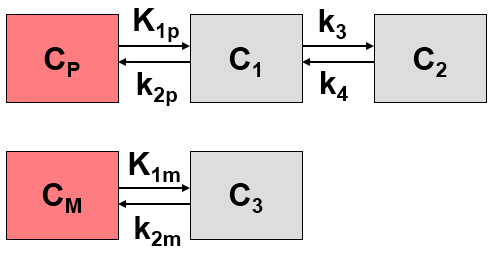
Differential Equation of the Mass Balance
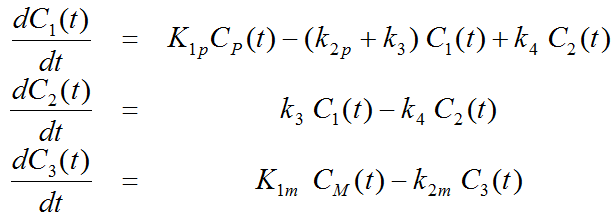
with the input curves of authentic ligand and metabolite, Cp(t) and CM(t), respectively. The input curve loading menu shows two corresponding entries as soon as the model is selected.
Operational Model Curve
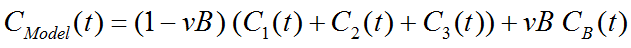
with the concentration CB(t) of tracer in whole blood, and the blood volume fraction vB.
Parameter Fitting using K1/k2 and VS
The 3-Tissue Compartments with Metabolites, K1/k2, Vs model variant uses the parameter ratios K1p/k2p, K1m/k2m, K1p/K1m, and Vs=K1p/k2p*k3/k4 as fitting parameters instead of k2p, K1m , k2m, and k4. The advantage of this formulation is that the ratios may be fixed at known or assumed values, or estimated in a coupled fit. The other three fitting parameters vB, K1p and k3 are the same as for the 3-Tissue Compartments with Metabolites model.
Reference
1.Fujita M, Seibyl JP, Verhoeff NP, Ichise M, Baldwin RM, Zoghbi SS, Burger C, Staley JK, Rajeevan N, Charney DS et al: Kinetic and equilibrium analyses of [(123)I]epidepride binding to striatal and extrastriatal dopamine D(2) receptors. Synapse 1999, 34(4):290-304.