Most of the parameters correspond to the parameters of the original ANTS software, for which a manual page is available. Please refer to the ANTS user community for hints regarding tailoring of the parameters for specific tasks.
Basic Parameters
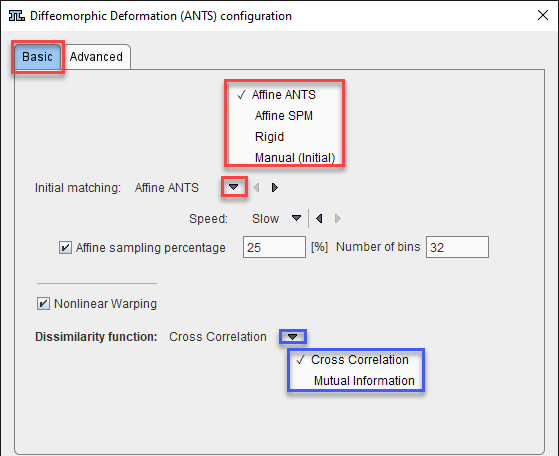
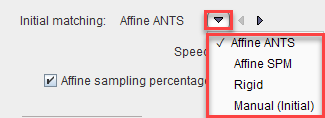
Initial Matching offers four methods to roughly align the data, before the elastic deformations start. The choices are
with Affine ANTS as defaults. It estimates and apply an affine transformation before the nonlinear warping iterations start. The amount of voxels used for calculation is specified by the sampling percentage parameter and represents a regular sampling strategy in combination with the histogram Number of bins. The initialization Speed has two options: Slow (![]() ) and Fast (
) and Fast (![]() ). The Slow Speed is more time consuming but the result are expected to be most accurate.
). The Slow Speed is more time consuming but the result are expected to be most accurate.
Affine SPM from Template-based Normalization (SPM5) represents the second choice.
Rigid will apply the standard Rigid Registration procedure, whereas Manual (Initial) will use the current location of the input image and not do any further alignment. The parameters of these methods (if applicable) appear after selecting them.
Nonlinear Warping should be enabled, otherwise the elastic part will be skipped. This can be helpful to assess the effect of the initial step.
The Dissimilarity function represents the definition of the matching algorithm. The available selections are:
•Cross correlation: to be considered when matching images with similar pixel distribution (same image modality). It is a time consuming calculation.
•Mutual information: to be considered when matching images with different pixel distribution (cross modality). It is a faster algorithm
Advanced Parameters
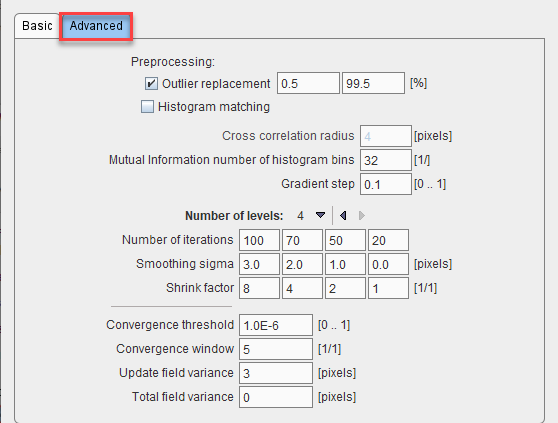
With Outlier replacement enabled, extreme pixel values are replaces by the winsorization method. Histogram matching normalizes the pixel values, and is recommended when matching images from the same modality.
The Histogram matching, when enabled, allows normalizing the pixels values by the histogram matching. It is a parameter to be used for matching images acquired using the same modality.
The Cross correlation radius determines the number of pixels in the neighborhood, which are used for calculating the cross correlation cost function.
The Gradient step characterizes the gradient descent optimization.
Number of levels determines the levels of hierarchical matching, working from coarse towards fine resolution. On each level, three parameters are configured: Shrink factor defines the sub-sampling in each direction. A factor of 8 reduces the number of pixels by a factor 83. Smoothing sigma is the Gaussian smoothing kernel size, and Number of iterations the number of optimizations at each level.
The Convergence parameters determine whether the iterations can be stopped before the configured number of iterations are exhausted. The Field variance parameters may be useful when adjusting to published methods.
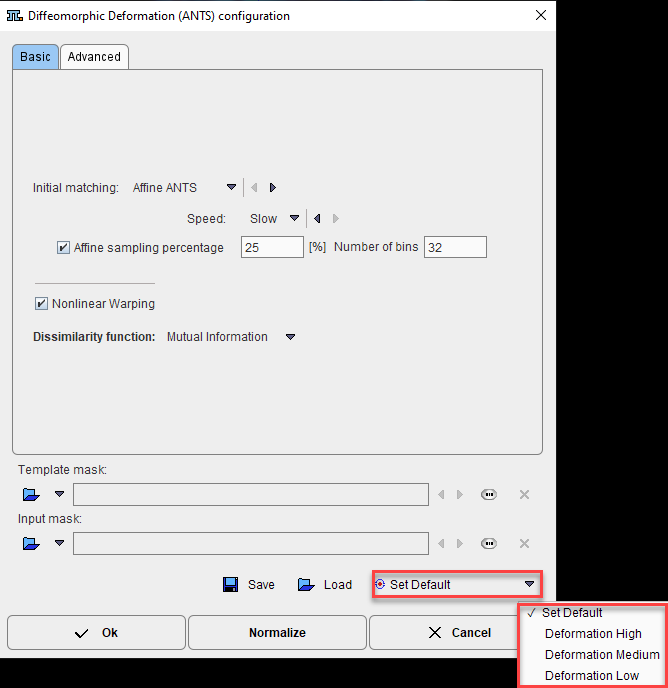
The Template mask allows defining a mask for the reference image if none was created and/or set to the matching protocol after the reference image was loaded. To discard the mask activate the Clear file or directory button ![]()
The Input mask allows defining a mask for the input image if none was created and/or set to the matching protocol after the input image was loaded. To discard the mask activate the Clear file or directory button ![]()
The Save icon allows saving the parameters set in the Basic and Advanced panel for later use.
The Load icon allows retrieving parameters previously saved and set them to the current matching procedure.
The Set Default options allows resetting the Basic and Advance parameters to their default values. The default values will depend on the selected entry in the default list. With Deformation High there will be long calculation times but with high quality deformation results while Deformation Low will return the results faster but the deformation quality will be lower. When hoovering with the mouse cursor above the selection, a tool tip will display the matching method (cross correlation=CC or mutual information=MI) and the associated default parameters. Please note that this parameters can be further modified on the Basic and/or Advanced panel and the new parameters saved using the Save icon.