Atlas Definition
VOI atlases can be easily prepared for use in PMOD. Three sets of data are required, the
1.Atlas images: They must be prepared as NifTI files and encode the atlas VOIs as numeric labels. Each pixel has a value of 0 if it is a background pixel, or is otherwise an integer number. We recommend using the HFS orientation (head first, supine = radiological convention) for human data.
2.Label list: A text file ending in .txt containing at least three columns. Each VOI is represented by a line of the form:
name1 label_name1 label_value1
name2 label_name2 label_value2
...
There must not be blank characters within the abbreviations or the names.
3.A manifest text file for the encoding of the species, for instance containing
SPECIES = HUMAN
The species supported are HUMAN, PRIMATE, PIG, RAT, MOUSE.
Note that the name of the atlas sub-directory in resources/templates/voitemplates must also be used for the template images, the label list, and the manifest. This name then shows up in the list of available atlases in the VOI analysis tool.
Included in the distribution are several templates. The naming and the arrangement of the files are illustrated below using the AAL-Merged atlas as an example.
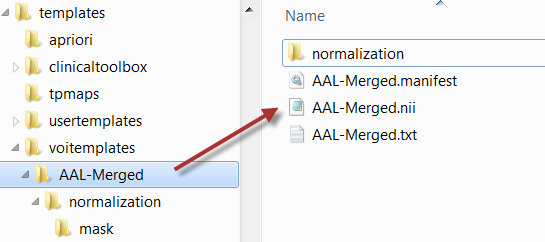
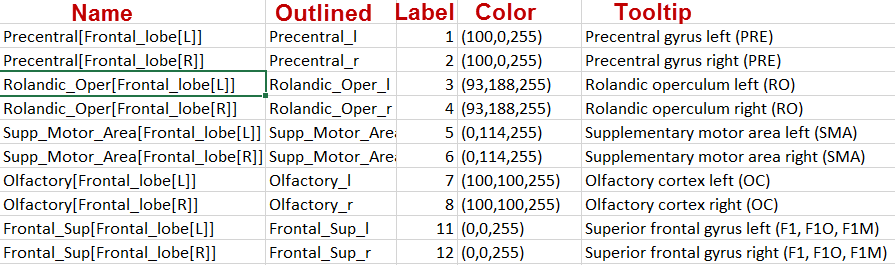
The AAL-Merged.txt contains the following 5 columns. The first column starts with the name followed by a bracket construction which encodes a tree structure. For instance, Precentral belongs to the Frontal_Lobe which is located in the L or right R hemisphere. The second column indicates the name of a generated contour VOI. The third column contains the label value in the atlas file. Each pixel in AAL-Merged.nii with value 1 will belong the Precentral_l VOI, pixels with value 2 to Precentral_l, etc.
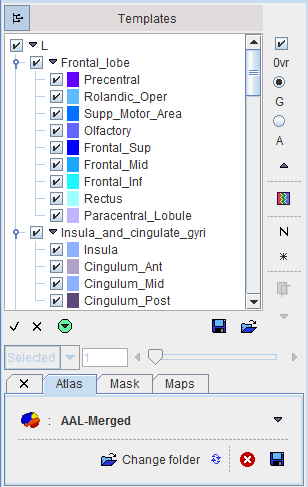
The corresponding VOI atlas structure is illustrated below.
Normalization Files
Atlases can only be applied to images if they have the same resolution and show the anatomy with the same geometry. Therefore, images originating from real experiments first need a normalization step for the atlas to be applied. This is done by calculating a normalization transform between the subject image and a "template" image representing the standard anatomy imaged with a certain modality, and using it for warping the VOIs to the subject anatomy.
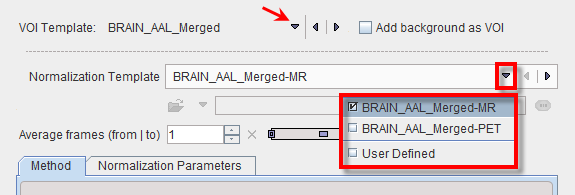
Appropriate template images need to be copied to a normalization sub-folder. In the example below normalization contains a T1 MR template with skull, as well as a PET template. Note that user defined templates names have to start with the atlas specific name, e.g. BRAIN_AAL_Merged, followed by the corresponding template modality -PET or -MRI. All of these templates show the anatomy in the space in which the BRAIN_AAL_Merged VOIs were defined.
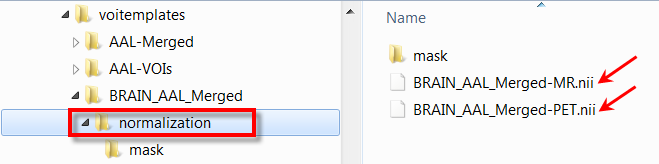
Correspondingly the Normalization Template list has the following entries.
The normalization works best if the information is restricted to the relevant image part. Therefore, normalization should contain a mask sub-folder with a mask file mask.nii containing 1 for all relevant pixels and 0 for all others.
Note: For atlases with SPECIES = HUMAN and SPACE = MNI in the manifest file the preparation of the normalization subdirectory is not required, as the universal files for the human brain are available in the template and the tpmaps directories.