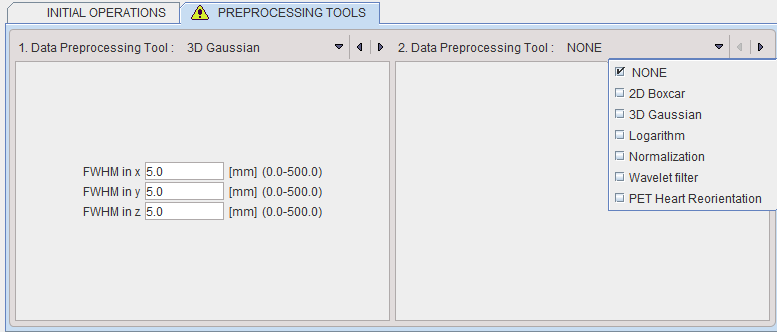
The tab PREPROCESSING TOOLS makes supports the application of two successive filters to the data.
Data Normalization
Normalization is a facility for transforming reconstructed image counts into activity concentration values calibrated in kBq/cc.
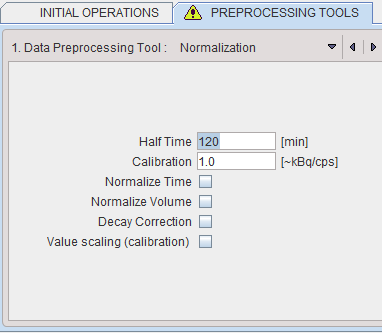
The following parameters are applicable during the calibration process:
▪Half Time is used for a correction of the physical decay to the acquisition start, if the Decay Correction box is enabled.
▪Calibration is a factor for the conversion of the measured counts - which represent only a fraction of the emitted photons - into the true number of physical decays. It is applied it the Value Scaling flag is enabled.
Note: The calibration factor can be determined using a phantom filled with a known, representative activity concentration. Phantom images are acquired, corrected and reconstructed using the same protocol as the research study. Then, the image values are decay corrected, time and volume normalized, resulting in images with counts per ml per second. As the next step, a homogeneous VOI is outlined and the average pixel value calculated. Finally, the calibration factor is calculated by dividing the known true phantom activity concentration by the VOI average.
▪Normalize Time is needed, if the pixel values represent total accumulated counts during the acquisition. If the box is checked, the image values are divided by the acquisition duration.
▪Normalize Volume is needed, if the pixel values represent activity, not activity concentration. If the box is checked, the values are divided by the image voxel size (known from the image header).
▪Decay Correction: If the box is checked, the pixel values are decay corrected to time zero. Note that under normal circumstances the start of the first frame will correspond to time zero, so decay correction is to the acquisition start. If this is not adequate, for instance in a PET image which is to be analyzed by an autoradiographic model, the frame start/end times should be edited (e.g. set to 40min and 50min for a 10-minute scan starting 40 minutes after injection), the image saved, and then loaded again for decay correcting to the injection time.
Wavelet filter
Wavelet is a time-domain filter for dynamic data which de-noises the time vector in each pixel separately.
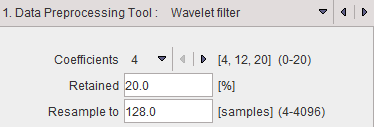
The wavelet filter uses the configured number (4, 12 or 20) of Daubechies Coefficients.
Retained specifies the percentage of frequencies retained before applying the inverse wavelet transformation. The smaller the percentage, the smoother the result.
Wavelet filtering requires a signal length of a power of 2. Therefore, in most cases signal re-sampling has to be performed to a 2n number specified in the Resample to field. Re-sampling uses linear interpolation.
Heart Reorientation
The PET Heart Reorientation is a facility that allows heart images re-orientation in short axis during loading. Please refer to the Cardiac PET Tool User Guide for the motivation of this tool.
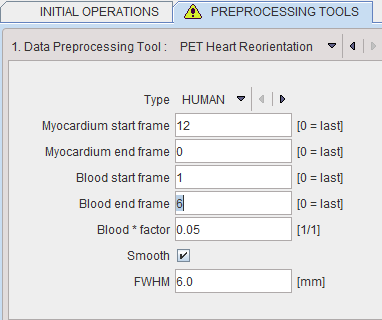
The following parameters need to be defined:
▪Type is representing the heart model type and can be selected from the available selection list: HUMAN, RAT or MOUSE according to the image to be analyzed.
▪Myocardium start frame and Myocardium end frame are used to define the frames average range for creating the myocardium averaged image.
▪Blood start frame and Blood end frame are used to define the frames average range for creating the blood averaged image.
▪Blood*factor: as there may exist some activity in the cavities, a fraction of the blood volume image can be subtracted to improve the contrast. In the example above a fraction of 0.05 of the blood averaged image will be subtracted.
▪Smooth and FWHM: optionally, the blood and myocardium averaged image can be smoothed with a 3D Gaussian filter with full-width half maximum value defined in the FWHM text box.
Note: The preprocessing tools are plug-ins and can be configured in the main configuration dialog.